
CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES ›› 2020, Vol. 38 ›› Issue (3): 310-316.doi: 10.12140/j.issn.1000-7423.2020.03.09
• ORIGINAL ARTICLES • Previous Articles Next Articles
ZHOU Hong-rang, MAO Guang-yao, WANG Xiao-ling, CHEN Mu-xin, YU Qing, WANG Ying, Ai Lin, XIAO Ning*( )
)
Received:2019-11-29
Online:2020-06-30
Published:2020-07-07
Contact:
Ning XIAO
E-mail:xiaoning@nipd.chinacdc.cn
Supported by:CLC Number:
ZHOU Hong-rang, MAO Guang-yao, WANG Xiao-ling, CHEN Mu-xin, YU Qing, WANG Ying, Ai Lin, XIAO Ning. Establishment and application of a multiplex recombinase-aided isothermal amplification technique for identifying Echinococcus granulosus and Echinococcus multilocularis[J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(3): 310-316.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.jsczz.cn/EN/10.12140/j.issn.1000-7423.2020.03.09

Fig. 1
Multiplex RAA and multiplex PCR amplifications using genomic DNA of Echinococcus granulosus G1 and E. multilocularis as templates A: Multiplex RAA amplification; B: Multiplex PCR amplification. M: DNA marker; 1: Echinococcus granulosus G1; 3: E. multilocularis; 5: E. granulosus G1 and E. multilocularis; 2, 4, 6: Negative control
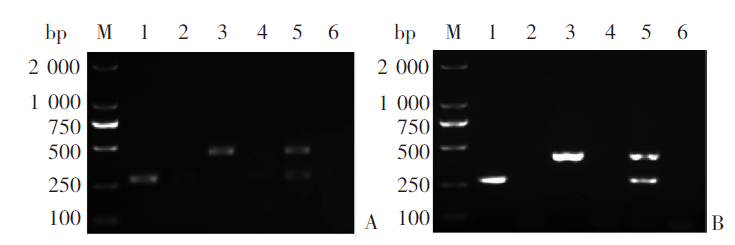
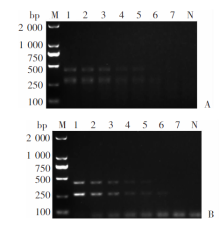
Fig. 3
Sensitivity of multiplex RAA assay in detecting Echinococcus granulosus G1 and E. multilocularis genomic DNA A: Multiplex RAA amplification; B: Multiplex PCR amplification. M: DNA marker; 1-7: Amplified products from 10.00, 5.00, 1.00, 0.50, 0.10, 0.05 and 0.01 ng/μl genomic DNA, respectively; N: Negative control
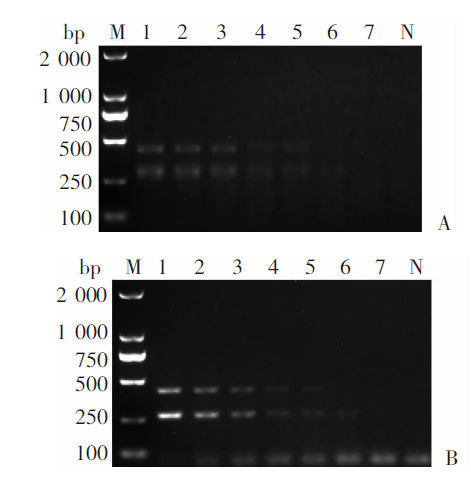

Fig. 5
Specificity of mRAA in detecting Echinococcus granulosus G1 and E. multilocularis genomic DNA A: Multiplex RAA amplification; B: Multiplex PCR amplification. M: DNA marker; 1: E. granulosus G1 and E. multilocularis; 2: E. granulosus G1; 3: E. multilocularis; 4: Taenia saginata; 5: T. asiatica; 6: T. multiceps; 7: Dipylidium caninum; 8: Toxocara canis; 9: Trichuris trichiura Linnaeus; 10: Giardia lamblia; 11: Fasciola hepatica; 12: Paragonimus westermani; 13: F. gigantica; 14: Clonorchis sinensis; N: Negative control
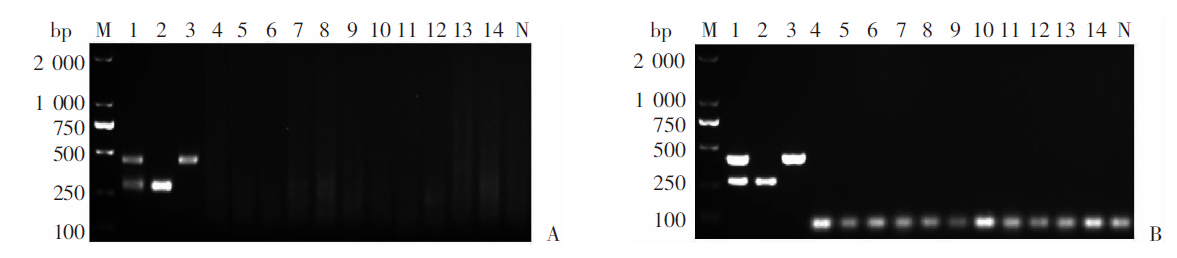
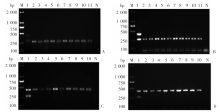
Fig. 6
Feasibility of mRAA method for liver tissue samples with Echinococcus infections A: Multiplex RAA detection of animal tissue samples infected with E. granulosus G1; B: Multiplex PCR detection of animal tissue samples infected with EgG1; C: Multiplex RAA detection of animal tissue samples infected with E. multilocularis; D: Multiplex PCR detection of animal tissue samples infected with Em. M: DNA marker; 1: Positive control; 2-9, 11: Animal tissue samples infected with Echinococcus granulosus G1; 10: Animal tissue samples infected with E. multilocularis; N: negative control
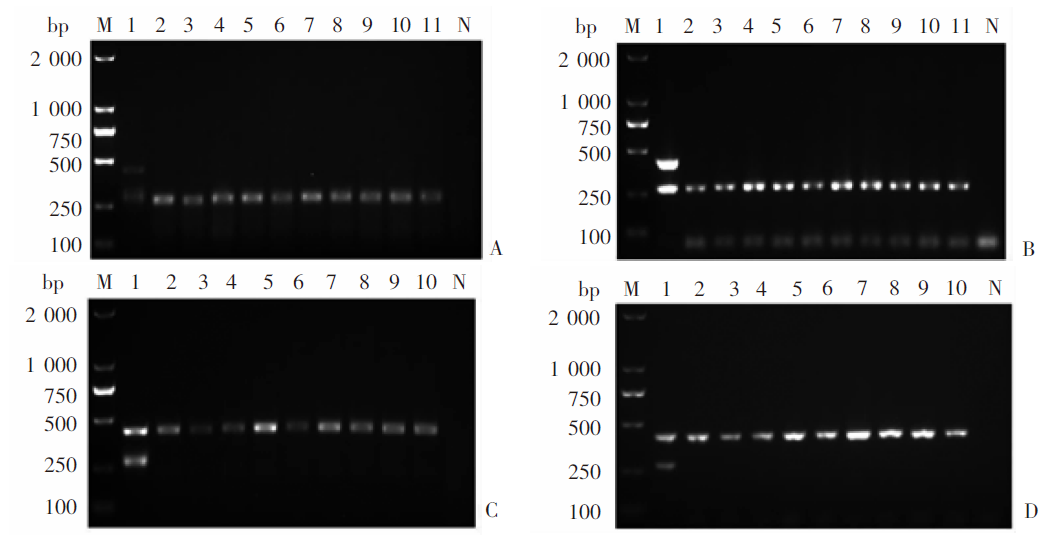
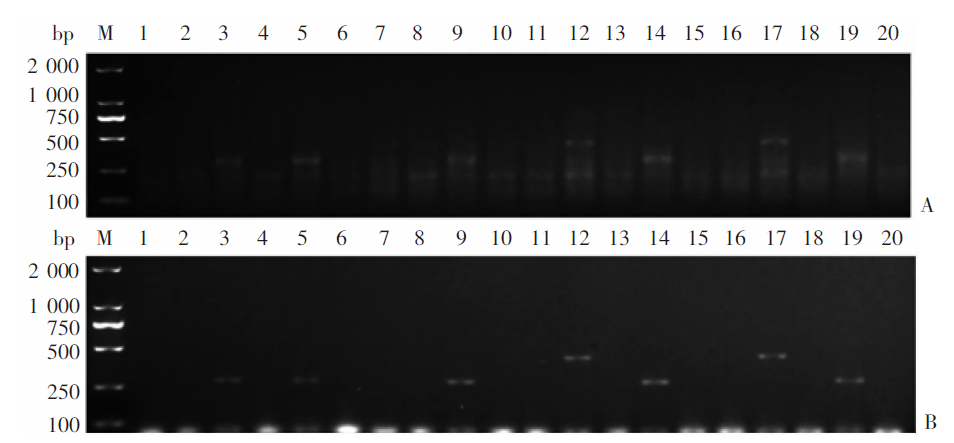
Fig. 8
Feasibility of the mRAA method for field fecal samples of dogs with Echinococcus infections A: Multiplex RAA; B: Multiplex PCR. M: DNA marker; 3, 5, 9, 14 and 19: Dog fecal samples positive for Echinococcus granulosus G1 infection; 12 and 17: Dog fecal samples positive for E. multilocularis infection; 1-2、4、6-8、10-11、13、15-16、18、20: Negative dog fecal samples
| [1] | Liu H, Xiao N, Yang SJ, et al. Epidemiological characteristics of canine Echinococcus infection in Qinghai-Tibet plateau of China[J]. Chin J Schisto Control, 2017,29(2):129-138. (in Chinese) |
| ( 刘辉, 肖宁, 杨诗杰, 等. 青藏高原地区犬棘球绦虫感染的流行病学特征[J]. 中国血吸虫病防治杂志, 2017,29(2):129-138.) | |
| [2] | Wu WP, Wang H, Wang Q, et al. A nationwide sampling survey on echinococcosis in China during 2012-2016[J]. Chin J Parasitol Parasit Dis, 2018,36(1):1-14. (in Chinese) |
| ( 伍卫平, 王虎, 王谦, 等. 2012-2016年中国棘球蚴病抽样调查分析[J]. 中国寄生虫学与寄生虫病杂志, 2018,36(1):1-14.) | |
| [3] |
Eckert J, Deplazes P. Biological, epidemiological, and clinical aspects of echinococcosis, a zoonosis of increasing concern[J]. Clin Microbiol Rev, 2004,17(1):107-135.
doi: 10.1128/cmr.17.1.107-135.2004 pmid: 14726458 |
| [4] | Wu WP. Prevalence and distribution of hydatidosis in China[J]. China Animal Heal, 2017,19(7):7-9. (in Chinese) |
| ( 伍卫平. 我国两型包虫病的流行与分布情况[J]. 中国动物保健, 2017,19(7):7-9.) | |
| [5] |
Enrico B, Peter K, Dominique AV. Expert consensus for the diagnosis and treatment of cystic and alveolar echinococcosis in humans[J]. Acta Trop, 2010,114(1):1-16.
doi: 10.1016/j.actatropica.2009.11.001 pmid: 19931502 |
| [6] |
Budke CM, Deplazes P, Torgerson PR. Global socioeconomic impact of cystic echinococcosis[J]. Emerging Infect Dis, 2006,12(2):296-303.
doi: 10.3201/eid1202.050499 pmid: 16494758 |
| [7] |
Torgerson PR, Keller K, Magnotta M, et al. The global burden of alveolar echinococcosis[J]. PLoS Negl Trop Dis, 2010,4(6):e722.
doi: 10.1371/journal.pntd.0000722 pmid: 20582310 |
| [8] | Zhang MY, Wu WP, Guan YY, et al. The analysis of disease burden of echinococcosis in China[J]. Chin J Parasitol Parasit Dis, 2018,36(1):15-19. (in Chinese) |
| ( 张梦媛, 伍卫平, 官亚宜, 等. 我国棘球蚴病疾病负担分析[J]. 中国寄生虫学与寄生虫病杂志, 2018,36(1):15-19.) | |
| [9] |
Karamon J. Detection of Echinococcus multilocularis in faeces by nested PCR with the use of diluted DNA samples[J]. Pol J Vet Sci, 2014,17(1):79-83.
doi: 10.2478/pjvs-2014-0010 pmid: 24724473 |
| [10] |
Dinkel A, Kern S, Brinker A, et al. A real-time multiplex-nested PCR system for coprological diagnosis of Echinococcus multilocularis and host species[J]. Parasitol Res, 2011,109(2):493-498.
doi: 10.1007/s00436-011-2272-0 |
| [11] | Knapp J, Millon L, Mouzon L, et al. Real time PCR to detect the environmental faecal contamination by Echinococcus multilocularis from red fox stools[J]. Vet Parasitol, 2014,201(1/2):40-47. |
| [12] | Trachsel D, Deplazes P, Mathis A. Identification of taeniid eggs in the faeces from carnivores based on multiplex PCR using targets in mitochondrial DNA[J]. Parasitology, 2007,134(6):911-920. |
| [13] |
Liu CN, Lou ZZ, Li L, et al. Discrimination between E. granulosus sensu stricto, E. multilocularis and E. shiquicus using a multiplex PCR assay[J]. PLoS Negl Trop Dis, 2015,9(9):e0004084.
doi: 10.1371/journal.pntd.0004084 pmid: 26393793 |
| [14] | Fan XX, Zhao YG, Li L, et al. Research progress of recombinant enzyme polymerase amplification (RPA) in rapid detection of diseases[J]. China Animal Heal Insp, 2016,33(8):72-77. (in Chinese) |
| ( 樊晓旭, 赵永刚, 李林, 等. 重组酶聚合酶扩增技术在疾病快速检测中的研究进展[J]. 中国动物检疫, 2016,33(8):72-77.) | |
| [15] |
Santiago-Felipe S, Tortajada-Genaro LA, Puchades R, et al. Recombinase polymerase and enzyme-linked immunosorbent assay as a DNA amplification-detection strategy for food analysis[J]. Anal Chim Acta, 2014,811:81-87.
doi: 10.1016/j.aca.2013.12.017 pmid: 24456598 |
| [16] |
Piepenburg O, Williams CH, Stemple DL, et al. DNA detection using recombination proteins[J]. PLoS Biol, 2006,4(7):e204.
doi: 10.1371/journal.pbio.0040204 pmid: 16756388 |
| [17] |
Abd El Wahed A, El-Deeb A, El-Tholoth M, et al. A portable reverse transcription recombinase polymerase amplification assay for rapid detection of foot-and-mouth disease virus[J]. PLoS One, 2013,8(8):e71642.
doi: 10.1371/journal.pone.0071642 pmid: 23977101 |
| [18] | LV B, Cheng HR, Yan QF, et al. Recombinase-aid amplification: a novel technology of in vitro rapid nucleic acid amplification[J]. Sci Sin Vitae, 2010,40(10):983-988. (in Chinese) |
| ( 吕蓓, 程海荣, 严庆丰, 等. 用重组酶介导扩增技术快速扩增核酸[J]. 中国科学: 生命科学, 2010,40(10):983-988.) | |
| [19] |
Li J, Macdonald J. Advances in isothermal amplification: novel strategies inspired by biological processes[J]. Biosens Bioelectron, 2015,64:196-211.
doi: 10.1016/j.bios.2014.08.069 pmid: 25218104 |
| [20] | Meng QH. Progress in the diagnosis and treatment of hydatidosis in China[J]. Chin J Urban Rural Enterp Hyg, 2014,29(5):16-18. (in Chinese) |
| ( 孟庆贺. 我国包虫病的诊断和治疗进展[J]. 中国城乡企业卫生, 2014,29(5):16-18.) | |
| [21] | Li YB, Yuan F, Li HW, et al. Application of fecal examination in investigation of infection source of echinococcosis[J]. J Ningxia Med Univ, 2010,32(1):147-150. (in Chinese) |
| ( 李燕兵, 袁芳, 李红卫, 等. 粪便检查在包虫病传染源调查中的应用[J]. 宁夏医科大学学报, 2010,32(1):147-150.) | |
| [22] |
Craig PS, Gasser RB, Parada L, et al. Diagnosis of canine echinococcosis: comparison of coproantigen and serum antibody tests with arecoline purgation in Uruguay[J]. Vet Parasitol, 1995,56(4):293-301.
doi: 10.1016/0304-4017(94)00680-b pmid: 7754606 |
| [23] |
Deplazes P, Alther P, Tanner I, et al. Echinococcus multilocularis coproantigen detection by enzyme-linked immunosorbent assay in fox, dog, and cat populations[J]. J Parasitol, 1999,85(1):115-121.
pmid: 10207375 |
| [24] | Wang SL, Wang LP, Wu LL, et al. Diagnostic value of nucleic acid detection in schistosomiasis japonica: a meta-analysis[J]. Chin J Schisto Control, 2020,32(1):15-22. (in Chinese) |
| ( 王盛琳, 王丽萍, 吴铃铃, 等. 核酸检测技术对日本血吸虫病诊断价值的meta分析[J]. 中国血吸虫病防治杂志, 2020,32(1):15-22.) | |
| [25] | Li T, Yang K. Application of isothermal amplification technology for pathogen detection in parasitic and other diseases[J]. Chin J Schisto Control, 2018,30(2):232-236. (in Chinese) |
| ( 李婷, 杨坤. 等温扩增技术在寄生虫及其他病原体检测中的应用[J]. 中国血吸虫病防治杂志, 2018,30(2):232-236.) | |
| [26] | Ni XW. Development and application of LAMP method for the identification of Echinococcus spp. in dog faeces[D]. Beijing: Chinese Academy of Agricultural Sciences, 2012. (in Chinese) |
| ( 倪兴维. 犬棘球绦虫粪LAMP检测方法的建立和应用[D]. 北京: 中国农业科学院, 2012.) | |
| [27] |
Wassermann M, Mackenstedt U, Romig T. A loop-mediated isothermal amplification (LAMP) method for the identification of species within the Echinococcus granulosus complex[J]. Vet Parasitol, 2014,200(1/2):97-103.
doi: 10.1016/j.vetpar.2013.12.012 |
| [28] |
Notomi T, Mori Y, Tomita N, et al. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects[J]. J Microbiol, 2015,53(1):1-5.
doi: 10.1007/s12275-015-4656-9 pmid: 25557475 |
| [29] | Luo LH, Zhang B. Loop-mediated isothermal amplification method for the detection of infectious diseases[J]. Chin J Front Heal Quar, 2014,37(1):68-72. (in Chinese) |
| ( 罗力涵, 张波. 环介导等温扩增技术及其在传染性疾病检测中的应用[J]. 中国国境卫生检疫杂志, 2014,37(1):68-72.) |
| [1] | CAO Deping, LI Jiajing, SONG Mengwei, MO Gang. Experimental observation on the changes of hepatic stellate cells stimulated in vitro with tissue protein of Echinococcus multilocularis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(4): 440-445. |
| [2] | DU Tao, HU Chunhui, GAN Xuehui, GAO Pan, ZHANG Fabin. Anti-Echinococcus multilocularis effect of total alkaloids of Sophora moorcroftiana in water solution and tablet forms in vitro and in vivo [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(1): 15-22. |
| [3] | HOU Xin-ling, LI De-wei, SHI Yang, WANG Mao-lin, ZIBIGU Rousu, ABIDAN Ainiwaer, ZHENG Xu-ran, KANG Xue-jiao, WANG Hui, LI Jing, ZHANG Chuan-shan. Changes of ST2+ T cell subset function and their immune checkpoint molecule expression in the peritoneal cavity of mice infected with Echinococcus multilocularis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(6): 708-716. |
| [4] | WU Liang-liang, YANG Ling-fei, SONG Tao. Ultrasound and pathological manifestations of lesions in SD rats with hepatic Echinococcus multilocularis infection established by different methods [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(4): 549-552. |
| [5] | ZHONG Shun-hu, SUN Yue, GUO Xiao-la, ZHENG Ya-dong, CHEN Yi-xia. Identification and bioinformatics analysis of differentially expressed miRNAs in splenic lymphocytes in Echinococcus multilocularis-infected mice [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(3): 288-294. |
| [6] | ZHUO Yi-cheng, YANG Hai-cheng, LIU Cheng-hao, ZHANG Bao-cai, DUO Xiao-yong, ZHANG Shi-jie. Effect of osteopontin expression level on the growth and development of Echinococcus multilocularis protoscoleces [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(3): 299-304. |
| [7] | ABUDUAINI Abulizi, PAIZULA Shalayiadang, TALAITI Tuergan, ZHANG Rui-qing, WANG Hui, ZHANG Chuan-shan, SHAO Ying-mei, TUERGANAILI Aji. Affect of Echinococcus multilocularis protein-mediated NK cell surface receptor NKG2A on the function of NK cells [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(1): 36-42. |
| [8] | HOU Jiao, WEN Hao, WANG Ming-kun, LI Wen-ding, LI liang, LI Jing, ZHANG Chuan-shan, WANG Hui. Changes of macrophage subsets and polarization in spleen of mice infected with Echinococcus multilocularis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(6): 771-778. |
| [9] | SHI Qi-qi, LIU Cong-shan, HUO Le-le, WEI Yu-fen, JIANG Bin, YIN Meng, XUE Jian, TAO Yi, ZHANG Hao-bing. Affect of aminoalcohol compound HT24 on the expression of tubulin in Echinococcus multilocularis protoscoleces [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(4): 437-443. |
| [10] | LI Ling-hui, WANG Wei, HOU Xin-ling, SHI Yang, LI De-wei, LI Liang, WANG Hui, LI Jing, ZHANG Chuan-shan. Affects of Echinococcus multilocularis metacestode infection on the natural killer T cells and their subsets in mouse spleen [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(3): 311-317. |
| [11] | GUO Bao-ping, GUO Gang, ZHANG Li, XIANG Jing-jing, WANG Xiao-ping, REN Yuan, QI Wen-jing, ZHANG Hui, LI Jun, ZHANG Wen-bao, WANG Hai-yan. Investigation on infection of Echinococcus multilocularis metacestode in small rodents in Chabchar County, Xinjiang [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(3): 327-332. |
| [12] | XU Kai, WANG Hai-jiu, ZHANG Li, ZHANG Yao-gang, FAN Hai-ning, REN Li, REN Bin, WANG Zhi-xin. Research progress on the mechanisms underlying the impairment of host hepatocytes by Echinococcus multilocularis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(2): 256-259. |
| [13] | ZHU Ji-hai, CAO De-ping, ZHAO Jun, LIU Jun, SHI Hu-xiang, LIU Yan. Investigation of differentially expressed genes in liver tissues of patients with alveolar echinococcosis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(1): 48-54. |
| [14] | ZHU Ji-hai, CAO De-ping, QIE Yangrangzhong, LIU Jun, ZHAO Jun, LIU Yan. Effect of Elsholtzia eriostachya in combination with albendazole in treatment of secondary Echinococcus multilocularis metacestode infection in rats [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(6): 688-694. |
| [15] | SHANG Jing-ye, ZHANG Guang-jia, YU Wen-jie, HE Wei, LIAO Sha, LI Rui-rui, HUANG Yan, LIU Yang, ZHONG Bo. Advances in researches on the genetic diversity of Echinococcus multilocularis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(5): 637-641. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||