
CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES ›› 2025, Vol. 43 ›› Issue (4): 518-525.doi: 10.12140/j.issn.1000-7423.2025.04.011
• ORIGINAL ARTICLES • Previous Articles Next Articles
LI Jianyong( )(
)( ), HE Biao, LI Meilin, LIU Taiping, ZHU Feng, ZHANG Jian, XU Wenyue*(
), HE Biao, LI Meilin, LIU Taiping, ZHU Feng, ZHANG Jian, XU Wenyue*( )(
)( )
)
Received:2025-01-24
Revised:2025-04-02
Online:2025-08-30
Published:2025-10-09
Contact:
E-mail: Supported by:CLC Number:
LI Jianyong, HE Biao, LI Meilin, LIU Taiping, ZHU Feng, ZHANG Jian, XU Wenyue. Rapid detection of mosquito species and their transmitted pathogens based on the CRISPR/Cas12a system[J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2025, 43(4): 518-525.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.jsczz.cn/EN/10.12140/j.issn.1000-7423.2025.04.011
Table 1
Primers of RPA, ssDNA and crRNA sequences
| 名称 Name | 序列(5′→3′) Sequence (5′→3′) |
|---|---|
| 斯氏按蚊RPA-F An. stephensi RPA-F | TTCATTACATTTAGCTGGAATTTCTTCAAT |
| 斯氏按蚊RPA-R An. stephensi RPA-R | ACGTAATTCCTGGCGATCGTATATTAATTA |
| 白纹伊蚊RPA-F Aedes albopictus RPA-F | CCCTTAATACTAGGAGCCCCTGATATAACT |
| 白纹伊蚊RPA-R Ae. albopictus RPA-R | CAGCAGTGTTAAAGAGGGGGGTAATATTCA |
| 约氏疟原虫BY265-RPA-F P. yoelii BY265-RPA-F | TTTTATATGTAGAAACTGCGAACGGCTCAT |
| 约氏疟原虫BY265-RPA-R P. yoelii BY265-RPA-R | CAAATACATGCTAATTACTCCGGAGAGTAA |
| Ⅰ型登革病毒RPA-F DENV1-RPA-F | ACCACCAGGGTACAGCTTCCCCTGGTGTTG |
| Ⅰ型登革病毒RPA-R DENV1-RPA-R | CCATGGAAGCTGTACGCATGGGGTAGCAGA |
| Ⅱ型登革病毒RPA-F DENV2-RPA-F | TCTGGTCTTTCCCAGCGTCAATATGCTGTT |
| Ⅱ型登革病毒RPA-R DENV2-RPA-R | TGAGATGAAGCTGTAGTCTCACTGGAAGGA |
| Ⅲ型登革病毒RPA-F DENV3-RPA-F | TCTGGTCTCTCCCAGCGTCAATATGCTGTT |
| Ⅲ型登革病毒RPA-R DENV3-RPA-R | ACTAGTGGTTAGAGGAGACCCCTCCCATGA |
| Ⅳ型登革病毒RPA-F DENV4-RPA-F | ATCTGATCGGAAAAGAGGAATATGTGGATT |
| Ⅳ型登革病毒RPA-R DENV4-RPA-R | AGCTACAGGCAGCACGGTTTGCTCAAGCCG |
| 单链DNA ssDNA | FAM-TTATT-BHQ1 |
| LF单链DNA LF-ssDNA | FAM-TTATT-Biotin |
| 斯氏按蚊crRNA An. stephensi crRNA | UAAUUUCUACUAAGUGUAGAUGGAGCAGUUAAUUUUAUUACU |
| 白纹伊蚊crRNA Ae. albopictus crRNA | UAAUUUCUACUAAGUGUAGAUCUCGAAUAAAUAAUAUAAGUU |
| 约氏疟原虫crRNA BY265 P. yoelii BY265 crRNA | UAAUUUCUACUAAGUGUAGAUUUAUAAGGAUAACUACGGAAA |
| Ⅰ型登革病毒crRNA DENV1 crRNA | UAAUUUCUACUAAGUGUAGAUGGAGGGGUCUCCUCUAACCAC |
| Ⅱ型登革病毒crRNA DENV2 crRNA | UAAUUUCUACUAAGUGUAGAUGGGGGGUCUCCUCUAACCUCU |
| Ⅲ型登革病毒crRNA DENV3 crRNA | UAAUUUCUACUAAGUGUAGAUCAGGAGGUACAGCUUCCCUCA |
| Ⅳ型登革病毒crRNA DENV4 crRNA | UAAUUUCUACUAAGUGUAGAUGAGAGUGAAGGAGUUCUGUAA |
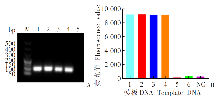
Fig. 1
Detection of cox1 gene in An. stephensi and Ae. albopictus using RPA (A) and CRISPR/Cas12a system (B) M: DNA marker; 1: An. stephensi larva; 2: An. stephensi adults; 3: Ae. albopictus larva; 4: Ae. albopictus adults; 5: Amplification of target fragment in Ae. albopictus using An. stephensi primers (A) and detection of Ae. albopictus using An. stephensi crRNA (B); 6: Detection of An. stephensi using Ae. albopictus crRNA; NC: Negative control.
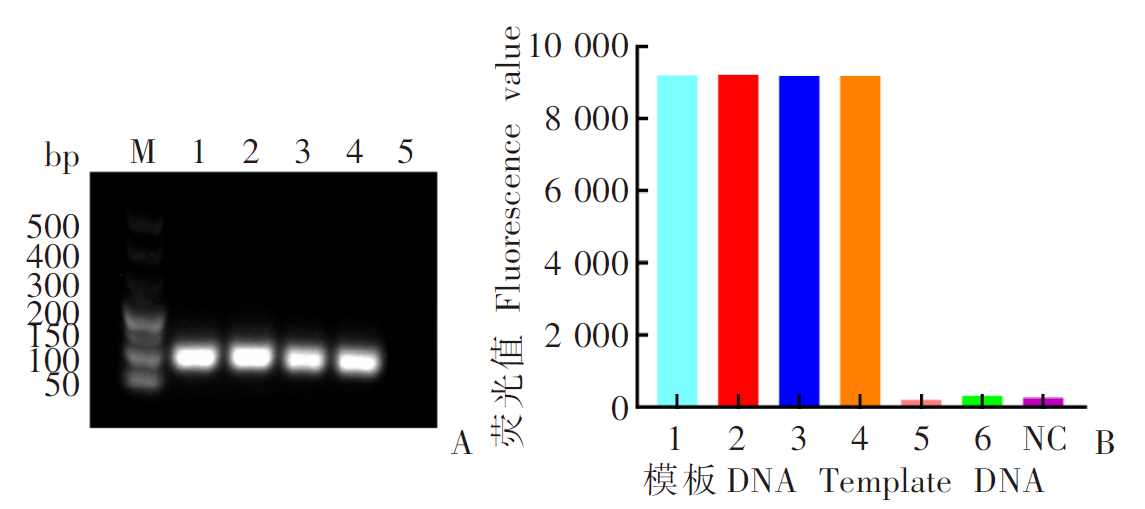

Fig. 4
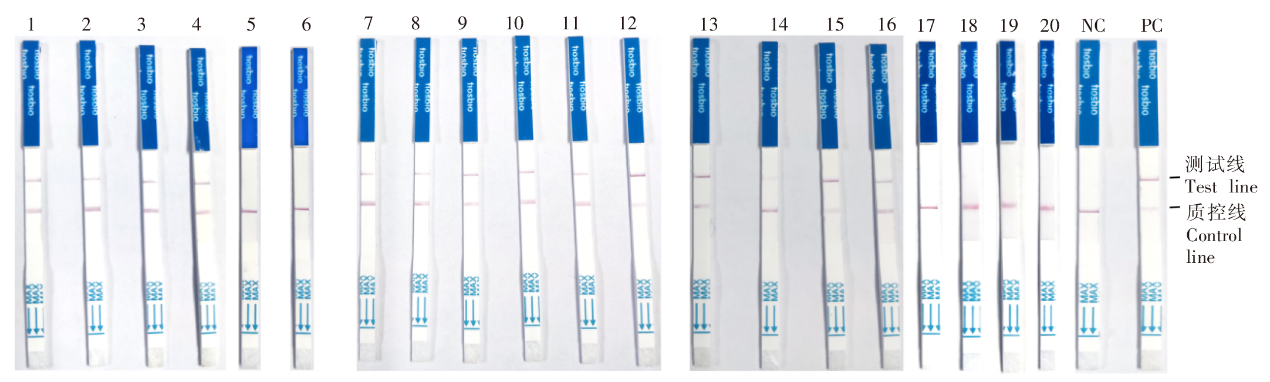
Detection of mosquito species, Plasmodium and dengue virus using test strips 1: An. stephensi larva; 2: An. stephensi adults; 3: Ae. albopictus larva; 4: Ae. albopictus adults; 5: Detection of Ae. albopictus using An. stephensi crRNA; 6: Detection of An. stephensi using Ae. albopictus crRNA; 7-12: Day 1, 2, 3, 4, 5, and 10 post-infection; 13: DENV1; 14: DENV2; 15: DENV3; 16: DENV4; 17: Detection of DENV2/3/4 using DENV1 crRNA; 18: Detection of DENV1/3/4 using DENV2 crRNA; 19: Detection of DENV1/2/4 using DENV3 crRNA; 20: Detection of DENV1/2/3 using DENV4 crRNA; NC: Negative control; PC: Positive control.
|
| [1] | WU Kai, XIA Qing, JIA Liming, FU Min. Research on development and application of an artifical intelligence platform assisting Plasmodium detection [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2025, 43(3): 345-350. |
| [2] | WANG Zhenyu, JIANG Li, YU Qing, ZHANG Yaoguang, WU Huanyu, CHEN Jian, ZHU Min. Interpretation of the Criteria for Detection of Malaria Parasite Nucleic Acid by Multiplex PCR Methods [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(4): 521-524. |
| [3] | WANG Rong, XU Jie, ZHU Xiaotong. Research advances on transmission-blocking vaccines targeting Plasmodium sexual stage [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(3): 399-406. |
| [4] | TAN Nie, JIAO Shiming, DING Yan, ZHU Chengyu, XU Wenyue. Effect of local complement activation in hepatocytes on the development of Plasmodium in the infrared phase [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(2): 169-176. |
| [5] | JIANG Ning, BAI Jie, LI Ping, SHAN Wenqi, ZHOU Qiuming, DONG Haowei, YUAN Hao, TAO Feng, LI Xiangyu, MA Yajun, PENG Heng. Establishment of a rapid detection method of mosquito-borne dengue virus based on loop-mediated isothermal amplification microfluidic chip technology [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(2): 234-241. |
| [6] | HAN Zhuxi, ZHU Xiaotong. Research progress on Plasmodium membrane protein complexes [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(1): 111-116. |
| [7] | LIANG Kejia, LIU Cong, LI Yanlin, LI Xiaoge, LIU Yan, LI Zhenkui. Research advances on transcriptional regulation in plasmodium sexual stages [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(5): 619-624. |
| [8] | SUN Jun. The biological significance of malarial hemozoin’s formation [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(2): 209-212. |
| [9] | GE Jie-yun, LIU Lei, SUN Yi-fan, CHENG Yang. Advances in research on the vacuolar membrane function and the associated proteins of plasmodium parasitophorous vacuole [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(3): 402-410. |
| [10] | LIU Jia, MA Jun-ying, ZHANG Xue-fei, WANG Hu, MA Xiao, CAI Hui-xia, GAO Qi, QIE Shuang, YIN Yan, WANG Wei, WANG Yong-shun, ZHANG Jing-xiao, LIU Yu-fang, LEI Wen, ZHAN Pei-zhen, ZHANG Qing. Evaluation of up-converting phosphor-lateral flow method for rapid detection of IgG antibodies in echinococcosis patients [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(1): 121-123. |
| [11] | LU Fei, ZHUO Xun-hui, LU Shao-hong. Research progress on the interaction between host cell autophagy and apicomplexa protozoa infection [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(6): 826-831. |
| [12] | LIU Hong, LIU Yao-bao, CAO Jun. Research advance and application of whole-genome sequencing of Plasmodium [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(2): 265-270. |
| [13] | LI Mei, TU Hong, XIA Zhi-gui, WANG Zhen-yu, ZHOU He-jun. Thermal stability of diagnostic targets Plasmodium falciparum histidine rich protein Ⅱ and Plasmodium lactate dehydrogenase in rapid detection [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(2): 245-248. |
| [14] | LUO Fei, TAN Yan, ZHOU Shuang, YUAN Yi, LI Shan-shan, XU Jing-ru, ZHOU Yang. Assessment and analysis of malaria diagnostic capacity by microscopy at primary health institutions in Chongqing [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(1): 93-99. |
| [15] | ZHU Ling-qian, FENG Xin-yu, HU Wei, LI Shi-zhu. Functions and roles of miRNA during the infection of Anopheles by Plasmodium [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(6): 742-748. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||