
CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES ›› 2024, Vol. 42 ›› Issue (5): 608-614.doi: 10.12140/j.issn.1000-7423.2024.05.007
• ORIGINAL ARTICLES • Previous Articles Next Articles
XU Guolei1( ), FENG Yanye2, HU Wei1,3,*(
), FENG Yanye2, HU Wei1,3,*( )
)
Received:2024-05-10
Revised:2024-07-21
Online:2024-10-30
Published:2024-10-17
Contact:
* E-mail: Supported by:CLC Number:
XU Guolei, FENG Yanye, HU Wei. Establishment and application evaluation of a rapid visualization detection method for Schistosoma japonicum specific nucleic acid fragments based on RPA-CRISPR/Cas12a technology[J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(5): 608-614.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.jsczz.cn/EN/10.12140/j.issn.1000-7423.2024.05.007
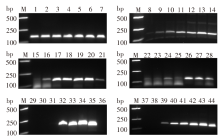
Fig. 1
Optimization of the RPA reaction M: DNA maker; 1-7: T4 recombinase Y concentrations of 30, 40, 50, 60, 70, 80, and 90 ng/μl, respectively; 8-14: T4 recombinase X concentrations of 40, 120, 200, 280, 360, 440, and 520 ng/μl, respectively; 15-21: T4 single-chain binding protein concentrations of 50, 100, 150, 200, 250, 300 and 350 ng/μl; 22-28: creatine kinase concentrations of 40, 60, 80, 100, 120, 140, and 160 ng/μl; 29-36: RPA reaction temperatures of 20, 25, 30, 35, 40, 45, 50 and 55 ℃, respectively; 37-44: RPA reaction times of 0, 5, 10, 15, 20, 30, 40 and 50 min, respectively.
| [1] | LoVerde PT. Schistosomiasis[M]. Adv Exp Med Biol, 2019, 1154: 45-70. |
| [2] | Basáñez MG, McCarthy JS, French MD, et al. A research agenda for helminth diseases of humans: modelling for control and elimination[J]. PLoS Negl Trop Dis, 2012, 6(4): e1548. |
| [3] | Xu J, Yang K, Li SZ, et al. Surveillance system after transmission control of schistosomiasis in P.R. China[J]. Chin J Schisto Control, 2014, 26(1): 1-5. (in Chinese) |
| (许静, 杨坤, 李石柱, 等. 我国血吸虫病传播控制后的监测体系[J]. 中国血吸虫病防治杂志, 2014, 26(1): 1-5.) | |
| [4] | Zhou XN. Implementation of precision control to achieve the goal of schistosomiasis elimination in China[J]. Chin J Schisto Control, 2016, 28(1): 1-4. (in Chinese) |
| (周晓农. 开展精准防治实现消除血吸虫病的目标[J]. 中国血吸虫病防治杂志, 2016, 28(1): 1-4.) | |
| [5] | Tong LB, Zhu CG, Lu K, et al. Evaluation on stool hatching method in detection of bovine schistosomiasis[J]. J Trop Dis Parasitol, 2012, 10(1): 8-10. (in Chinese) |
| (童来保, 朱传刚, 陆珂, 等. 粪便毛蚴孵化法检测牛血吸虫病的评价[J]. 热带病与寄生虫学, 2012, 10(1): 8-10.) | |
| [6] | Yao YY, Xiong CR, Dong X, et al. Application value of a dynamic automatic identification system in Schistosoma japonicum miracidium hatching test[J]. Chin J Schisto Control, 2019, 31(6): 652-654, 668. (in Chinese) |
| (姚韵怡, 熊春蓉, 董萱, 等. 血吸虫毛蚴动态自动识别系统在尼龙绢集卵孵化法中的应用价值[J]. 中国血吸虫病防治杂志, 2019, 31(6): 652-654, 668.) | |
| [7] | Zhang XJ, Cheng TY, Lu FZ, et al. Study on fast hatching test for determination of Schistosoma japonicum miracidium in stool[J]. ACTA Agric Zhejiangensis, 2004, 16(6): 381-383. (in Chinese) |
| (张雪娟, 程天印, 卢福庄, 等. 日本血吸虫病快速粪便毛蚴孵化法检测研究[J]. 浙江农业学报, 2004, 16(6): 381-383.) | |
| [8] | Xu R, Zhao DY, Lu K, et al. Development of colloidal gold immunochromatography assay strip for schistosomiasis diagnosis in domestic animals[J]. Chin J Schisto Control, 2015, 27(5): 474-478. (in Chinese) |
| (许瑞, 赵登云, 陆珂, 等. 诊断家畜日本血吸虫感染胶体金免疫层析试纸条的研制[J]. 中国血吸虫病防治杂志, 2015, 27(5): 474-478.) | |
| [9] | Chen YP, Weng XH, Shen XF, et al. Experimental study on detection of Schistosoma japonicum 5D gene by using PCR[J]. Chin J Parasitol Parasit Dis, 1998, 16(1): 58-61. (in Chinese) |
| (陈一平, 翁心华, 沈雪芳, 等. 聚合酶链反应检测日本血吸虫5D基因的实验研究[J]. 中国寄生虫学与寄生虫病杂志, 1998, 16(1): 58-61.) | |
| [10] | Lu ZX, Xu J, Gong W, et al. Detection of Schistosoma japonicum DNA by polymerase chain reaction[J]. Chin J Zoonoses, 2007, 23(5): 479-483. (in Chinese) |
| (陆正贤, 许静, 龚唯, 等. 聚合酶链反应检测日本血吸虫DNA的实验研究[J]. 中国人兽共患病学报, 2007, 23(5): 479-483.) | |
| [11] | Zeng FS, He L, He XM, et al. Nested-PCR assay for detection of Schistosoma japonicum[J]. J Trop Dis Parasitol, 2017, 15(3): 136-138. (in Chinese) |
| (曾凡胜, 何露, 何雪梅, 等. 日本血吸虫巢式PCR检测方法的建立[J]. 热带病与寄生虫学, 2017, 15(3): 136-138.) | |
| [12] |
Zhang X, He CC, Liu JM, et al. Nested-PCR assay for detection of Schistosoma japonicum infection in domestic animals[J]. Infect Dis Poverty, 2017, 6(1): 86.
doi: 10.1186/s40249-017-0298-y pmid: 28407808 |
| [13] | Zhang F, Hou M, Yu CX, et al. The use of nested PCR to monitor sentinel mice in aquatic areas infested with Schistosoma japonicum[J]. J Pathog Biol, 2015, 10(4): 325-328. (in Chinese) |
| (张凡, 侯敏, 余传信, 等. 巢式PCR法在日本血吸虫易感水域哨鼠监测中的应用[J]. 中国病原生物学杂志, 2015, 10(4): 325-328.) | |
| [14] | Liu AP, Yang QL, Guo JJ, et al. DNA detection of low-intensity Schistosoma japonicum infection by nested-PCR assay in serum of host[J]. Suzhou Univ J Med Sci, 2010, 30(5): 915-917, 930. (in Chinese) |
| (刘爱平, 杨巧林, 郭俊杰, 等. 巢式PCR法检测日本血吸虫低感染度宿主血清DNA的研究[J]. 苏州大学学报(医学版), 2010, 30(5): 915-917, 930.) | |
| [15] | Xu J, Duan ZL, Guan ZX, et al. Early detection of circulating DNA of Schistosoma japonicum in sentinel mice models[J]. Exp Parasitol, 2017, 176: 82-88. |
| [16] | Zhou L, Tang JF, Zhao YY, et al. A highly sensitive TaqMan real-time PCR assay for early detection of Schistosoma species[J]. Acta Trop, 2011, 120(1/2): 88-94. |
| [17] | Lier T, Simonsen GS, Haaheim H, et al. Novel real-time PCR for detection of Schistosoma japonicum in stool[J]. Southeast Asian J Trop Med Public Health, 2006, 37(2): 257-264. |
| [18] | Lier T, Simonsen GS, Wang TP, et al. Real-time polymerase chain reaction for detection of low-intensity Schistosoma japonicum infections in China[J]. Am J Trop Med Hyg, 2009, 81(3): 428-432. |
| [19] |
Xu J, Rong R, Zhang HQ, et al. Sensitive and rapid detection of Schistosoma japonicum DNA by loop-mediated isothermal amplification (LAMP)[J]. Int J Parasitol, 2010, 40(3): 327-331.
doi: 10.1016/j.ijpara.2009.08.010 pmid: 19735662 |
| [20] | Fernández-Soto P, Gandasegui Arahuetes J, Sánchez Hernández A, et al. A loop-mediated isothermal amplification (LAMP) assay for early detection of Schistosoma mansoni in stool samples: a diagnostic approach in a murine model[J]. PLoS Negl Trop Dis, 2014, 8(9): e3126. |
| [21] | Abbasi I, King CH, Muchiri EM, et al. Detection of Schistosoma mansoni and Schistosoma haematobium DNA by loop-mediated isothermal amplification: identification of infected snails from early prepatency[J]. Am J Trop Med Hyg, 2010, 83(2): 427-432. |
| [22] |
Weerakoon KG, Gordon CA, Williams GM, et al. Droplet digital PCR diagnosis of human schistosomiasis: parasite cell-free DNA detection in diverse clinical samples[J]. J Infect Dis, 2017, 216(12): 1611-1622.
doi: 10.1093/infdis/jix521 pmid: 29029307 |
| [23] | Cai PF, Weerakoon KG, Mu Y, et al. Comparison of Kato-Katz, antibody-based ELISA and droplet digital PCR diagnosis of schistosomiasis japonica: lessons learnt from a setting of low infection intensity[J]. PLoSNegl Trop Dis, 2019, 13(3): e0007228. |
| [24] | Piepenburg O, Williams CH, Stemple DL, et al. DNA detection using recombination proteins[J]. PLoS Biol, 2006, 4(7): e204. |
| [25] | Fan XX, Zhao YG, Li L, et al. Research progress of recombinant enzyme polymerase amplification(RPA) in rapid detection of diseases[J]. China Anim Health Insp, 2016, 33(8): 72-77. (in Chinese) |
| (樊晓旭, 赵永刚, 李林, 等. 重组酶聚合酶扩增技术在疾病快速检测中的研究进展[J]. 中国动物检疫, 2016, 33(8): 72-77.) | |
| [26] | East-Seletsky A, O’Connell MR, Knight SC, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection[J]. Nature, 2016, 538(7624): 270-273. |
| [27] | Abudayyeh OO, Gootenberg JS, Konermann S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector[J]. Science, 2016, 353(6299): aaf5573. |
| [28] | The Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium. The Schistosoma japonicum genome reveals features of host-parasite interplay[J]. Nature, 2009, 460(7253): 345-351. |
| [29] |
Guo QH, Chen C, Zhou KK, et al. Evaluation of a real-time PCR assay for diagnosis of schistosomiasis japonica in the domestic goat[J]. Parasit Vectors, 2020, 13(1): 535.
doi: 10.1186/s13071-020-04420-8 pmid: 33109260 |
| [30] | Sun K, Xing WW, Yu XL, et al. Recombinase polymerase amplification combined with a lateral flow dipstick for rapid and visual detection of Schistosoma japonicum[J]. Parasit Vectors, 2016, 9(1): 476. |
| [31] | Xing WW, Yu XL, Feng JT, et al. Field evaluation of a recombinase polymerase amplification assay for the diagnosis of Schistosoma japonicum infection in Hunan Province of China[J]. BMC Infect Dis, 2017, 17(1): 164. |
| [32] | Deng WP, Xu B, Hong QH, et al. Establishment of the detection method for Schistosoma japonicum by recombinase polymerase amplification combined with electrochemical DNA biosensor[J]. Chin J Parasitol Parasit Dis, 2020, 38(2): 168-174. (in Chinese) |
|
(邓王平, 徐斌, 洪清华, 等. 重组酶聚合酶扩增结合电化学DNA传感器检测日本血吸虫方法的建立[J]. 中国寄生虫学与寄生虫病杂志, 2020, 38(2): 168-174.)
doi: 10.12140/j.issn.1000-7423.2020.02.006 |
|
| [33] | Deng WP, Hong QH, Xu B, et al. Development and preliminary evaluation of a rapid visualization detection method for circulating nucleic acids of Schistosoma japonicum based on RPA-LFD[J]. Chin J Parasitol Parasit Dis, 2020, 38(3): 286-292. (in Chinese) |
|
(邓王平, 洪清华, 徐斌, 等. 基于RPA-LFD的日本血吸虫循环核酸快速可视化检测方法的建立及初步评价[J]. 中国寄生虫学与寄生虫病杂志, 2020, 38(3): 286-292.)
doi: 10.12140/j.issn.1000-7423.2020.03.005 |
|
| [34] | Guo QH, Zhou KR, Chen C, et al. Development of a recombinase polymerase amplification assay for schistosomiasis japonica diagnosis in the experimental mice and domestic goats[J]. Front Cell Infect Microbiol, 2021, 11: 791997. |
| [35] | Zhao S, Li T, Yang K, et al. Establishment of a recombinase-aided isothermal amplification technique to detect Schistosoma japonicum specific gene fragments[J]. Chin J Schisto Control, 2018, 30(3): 273-277, 306. (in Chinese) |
| (赵松, 李婷, 杨坤, 等. 重组酶介导的日本血吸虫特异性基因片段核酸等温扩增检测方法的建立[J]. 中国血吸虫病防治杂志, 2018, 30(3): 273-277, 306.) | |
| [36] | Zhao S, Liu YH, Li T, et al. Rapid detection of Schistosoma japonicum specific gene fragment by recombinase aided isothermal amplification combined with fluorescent probe[J]. Chin J Parasitol Parasit Dis, 2019, 37(1): 23-27. (in Chinese) |
|
(赵松, 刘燕红, 李婷, 等. 结合重组酶介导的核酸等温扩增和荧光探针快速检测日本血吸虫基因片段[J]. 中国寄生虫学与寄生虫病杂志, 2019, 37(1): 23-27.)
doi: 10.12140/j.issn.1000-7423.2019.01.005 |
|
| [37] | Li T, Liu YH, Zhao S, et al. Rapid detection of Schistosoma japonicum-infected snails with recombinaseaided isothermal amplification assay[J]. Chin J Schisto Control, 2019, 31(2): 109-114, 120. (in Chinese) |
| (李婷, 刘燕红, 赵松, 等. 重组酶介导的核酸等温扩增荧光法快速检测日本血吸虫感染性钉螺[J]. 中国血吸虫病防治杂志, 2019, 31(2): 109-114, 120.) | |
| [38] | Ye YY, Zhao S, Liu YH, et al. Establishment of a nucleic acid dipstick test for detection of Schistosoma japonicum specific gene fragments based on the recombinase-aided isothermal amplification assay[J]. Chin J Schisto Control, 2021, 33(4): 334-338, 395. (in Chinese) |
| (叶钰滢, 赵松, 刘燕红, 等. 基于重组酶介导核酸等温扩增技术的日本血吸虫特异性基因片段核酸试纸条检测方法的建立[J]. 中国血吸虫病防治杂志, 2021, 33(4): 334-338, 395.) | |
| [39] | Liu JY, Wang XX, Sheng F, et al. Metagenomic sequencing for identifying pathogen-specific circulating DNAs and development of diagnostic methods for schistosomiasis[J]. iScience, 2023, 26(9): 107495. |
| [1] | ZHANG Lesheng, WANG Qi, WANG Fengfeng, ZHU Hai, LI Qingyue, MA Xiaohe, WANG Min, WANG Yujie, WANG Tianping, CAO Zhiguo. Establishment of multienzyme isothermal rapid amplification assay combined with fluorescent probe for rapid detection of Schistosoma japonicum gene [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(4): 481-486. |
| [2] | WANG Ning, PENG Hanqi, GAO Changzhe, CHENG Yuheng, LYU Dabing. Characterization of the mitochondrial genome and phylogenetic implication of Schistosoma japonicum featured with “nocturnal cercarial emergence” [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(6): 699-707. |
| [3] | TAN Xiao, ZHU Qi, LIU Zhongqi, LI Jia, PENG Dingjin. Immunogenicity of Schistosoma japonicum Sj26gst mRNA vaccine candidate [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(5): 546-551. |
| [4] | LIU Huaman, Bikash Giri, FANG Chuantao, ZHENG Yameng, WU Huixin, ZENG Minhao, LI Shan, CHENG Guofeng. Identification of gender associated m6A modified circRNA in Schistosoma japonicum [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(5): 552-558. |
| [5] | XU Yin, LIU Ting, XU Hui, ZENG Xiaojun, LAN Weiming, GONG Zhihong, DAI Kunjiao, QIU Tingting, HAO Xian, XIE Shuying. Establishment and application of PCR-CRISPR/Cas12a-based detection method for Clonorchis sinensis metacercaria [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(4): 421-426. |
| [6] | LAN Weiming, XU Hui, XU Yin, QIU Tingting, XIE Shuying, DENG Fenglin, HU Shaoliang, LIU Huan, GUO Jiagang, ZENG Xiaojun. Study on early warning of high risk environment of Schistosoma japonicum infection by quantitative real-time PCR [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(4): 502-505. |
| [7] | WANG Xiao-ling, ZHANG Wei, YI Cun, CHEN Xiang-yu, YANG Wen-bin, XU Bin, HU Wei. The effect of SjGPR89 protein on the growth and development of Schistosoma japonicum [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(6): 701-707. |
| [8] | CHEN Guo, ZHU Dan-dan, DUAN Yi-nong. Research progress of immune regulation protein B7 family on immune regulation during Schistosoma japonicum infection [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(6): 774-779. |
| [9] | YAN Xiao-lan, WEN Li-yong, XIONG Yan-hong, ZHENG Bin, ZHANG Jian-feng, WANG Tian-ping, YU Li-ling, XU Guo-zhang, LIN Dan-dan, ZHOU Xiao-nong. Interpretation of Criteria for Detection of Antibody against Schistosoma japonicum—Enzyme-linked Immunosorbent Assay [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(6): 798-800. |
| [10] | TANG Xian-shi, JI Wen-xiang, XIONG Chun-rong, ZHOU Yong-hua, XU Yong-liang, TONG De-sheng. Study on anxiety-like behavior of mice with late-stage infection of Schistosoma japonicum [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(5): 622-628. |
| [11] | LIANG Le, ZHANG Jing, SHEN Yu-juan, HU Yuan, CAO Jian-ping. Cyclic guanosine monophosphate-adenosine monophosphate promotes liver egg granuloma formation and fibrosis in mice infected with Schistosoma japonicum [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(4): 441-445. |
| [12] | GAO Yuan, ZHANG Xiao-cheng, HU Yuan, CAO Jian-ping. Study on the inhibitory effect of natural killer cells on liver fibrosis of schistosomiasis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(2): 168-174. |
| [13] | HONG Yang, LIN Jiao-jiao. Research progress on proteomics in Schistosoma japonicum [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(6): 725-730. |
| [14] | HUANG Ai-long, ZHANG Bei, SHEN Han-yu, CHEN Guo, LI Jing, ZHU Dan-dan, DUAN Yi-nong. Expression and function of triggering receptor expressed on myeloid cells 1 in the liver of mice infected with Schistosoma japonicum [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(5): 621-626. |
| [15] | ZHAO Cheng-si, QIN Min, TAN Ming-juan, MIAO Ting-ting, SHAO Tian-ye, LIU Xin-jian, WANG Yong. Effect of praziquantel on impaired renal function in mice with acute infection of Schistosoma japonicum [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(2): 200-209. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||