
CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES ›› 2023, Vol. 41 ›› Issue (4): 421-426.doi: 10.12140/j.issn.1000-7423.2023.04.004
• ORIGINAL ARTICLES • Previous Articles Next Articles
XU Yin1( ), LIU Ting2, XU Hui1, ZENG Xiaojun1, LAN Weiming1, GONG Zhihong1, DAI Kunjiao1, QIU Tingting1, HAO Xian2, XIE Shuying1,*(
), LIU Ting2, XU Hui1, ZENG Xiaojun1, LAN Weiming1, GONG Zhihong1, DAI Kunjiao1, QIU Tingting1, HAO Xian2, XIE Shuying1,*( )
)
Received:2022-11-10
Revised:2023-01-31
Online:2023-08-30
Published:2023-09-06
Contact:
*E-mail: Supported by:CLC Number:
XU Yin, LIU Ting, XU Hui, ZENG Xiaojun, LAN Weiming, GONG Zhihong, DAI Kunjiao, QIU Tingting, HAO Xian, XIE Shuying. Establishment and application of PCR-CRISPR/Cas12a-based detection method for Clonorchis sinensis metacercaria[J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(4): 421-426.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.jsczz.cn/EN/10.12140/j.issn.1000-7423.2023.04.004
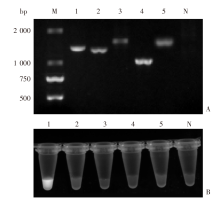
Fig. 4
Specificity analysis of the PCR-CRISPR/Cas12a system A: Agarose gel electrophoresis analysis of PCR amplicons from different parasite species; B: Results of PCR-CRISPR/Cas12a detection of different species of parasites. M: DNA marker; 1: Clonorchis sinensis; 2: Metorchis orientalis; 3: Spirometra erinaceieuropaei; 4: Anisakis pegreffii; 5: Taenia asiatica; N: Negative control.
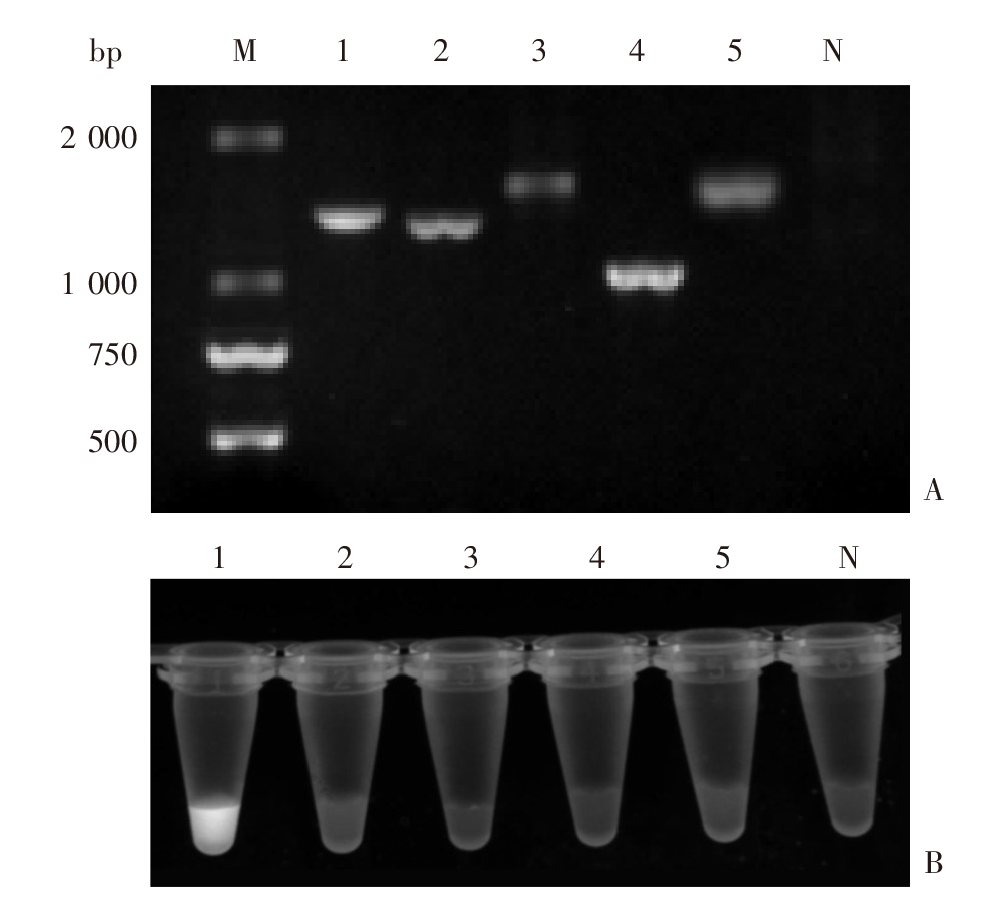

Fig. 5
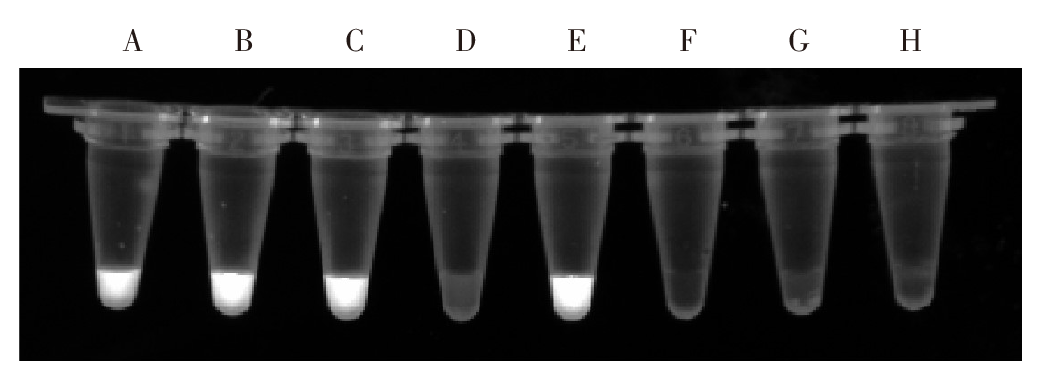
The results of PCR-CRISPR/Cas12a detection of fish flesh samples infected with the different species of metacercariae A: The fish sample infected with the metacercaria of C. sinensis, M. orientalis and unknown species; B: The fish sample infected with the metacercaria of C. sinensis and M. orientalis; C: The fish sample infected with the metacercaria of C. sinensis and unknown species; D: The fish sample infected with the metacercaria of M. orientalis and unknown species; E: The fish sample infected with the metacercaria of C. sinensis; F: The fish sample infected with the metacercaria of M. orientalis; G: The fish sample infected with the metacercaria of unknown species; H: Negative control.
| [1] |
Qian MB, Utzinger J, Keiser J, et al. Clonorchiasis[J]. Lancet, 2016, 387(10020): 800-810.
doi: 10.1016/S0140-6736(15)60313-0 |
| [2] |
Na BK, Pak JH, Hong SJ. Clonorchis sinensis and clonorchiasis[J]. Acta Trop, 2020, 203: 105309.
doi: 10.1016/j.actatropica.2019.105309 |
| [3] | Li X, Wang JC, Guan L, et al. Advances in the pathogenesis of cholangiocarcinoma caused by Clonorchis sinensis[J]. Chin J Parasitol Parasit Dis, 2020, 38(2): 250-254. (in Chinese) |
| (李潇, 王江畅, 管磊, 等. 华支睾吸虫致胆管癌发生发展的研究进展[J]. 中国寄生虫学与寄生虫病杂志, 2020, 38(2):250-254.) | |
| [4] |
Bouvard V, Baan R, Straif K, et al. A review of human carcinogens: Part B: biological agents[J]. Lancet Oncol, 2009, 10(4): 321-322.
doi: 10.1016/S1470-2045(09)70096-8 |
| [5] | Sohn WM. Fish-borne zoonotic trematode metacercariae in the Republic of Korea[J]. Korean J Parasitol, 2009, 47(Suppl): S103-S113. |
| [6] | Jing WW, Cheng XJ. Application and prospect of multidisciplinary new detection technology in the diagnosis of parasite infections[J]. Chin J Parasitol Parasit Dis, 2022, 40(1): 20-27, 35. (in Chinese) |
| (荆雯雯, 程训佳. 多学科交叉新型检测技术在寄生虫感染诊断中的应用和展望[J]. 中国寄生虫学与寄生虫病杂志, 2022, 40(1): 20-27, 35.) | |
| [7] |
Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes[J]. Science, 2007, 315(5819): 1709-1712.
doi: 10.1126/science.1138140 pmid: 17379808 |
| [8] |
Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system[J]. Cell, 2015, 163(3): 759-771.
doi: 10.1016/j.cell.2015.09.038 pmid: 26422227 |
| [9] |
Chen JS, Ma EB, Harrington LB, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity[J]. Science, 2018, 360(6387): 436-439.
doi: 10.1126/science.aar6245 pmid: 29449511 |
| [10] |
Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2[J]. Science, 2017, 356(6336): 438-442.
doi: 10.1126/science.aam9321 pmid: 28408723 |
| [11] |
Dueñas E, Nakamoto JA, Cabrera-Sosa L, et al. Novel CRISPR-based detection of Leishmaniaspecies[J]. Front Microbiol, 2022, 13: 958693.
doi: 10.3389/fmicb.2022.958693 |
| [12] |
Yu FC, Zhang KH, Wang YL, et al. CRISPR/Cas12a-based on-site diagnostics of Cryptosporidium parvum Ⅱd-subtype-family from human and cattle fecal samples[J]. Parasit Vectors, 2021, 14(1): 208.
doi: 10.1186/s13071-021-04709-2 |
| [13] |
Ma QN, Wang M, Zheng LB, et al. RAA-Cas12a-Tg: a nucleic acid detection system for Toxoplasma gondii based on CRISPR-Cas12a combined with recombinase-aided amplification(RAA)[J]. Microorganisms, 2021, 9(8): 1644.
doi: 10.3390/microorganisms9081644 |
| [14] |
Luton K, Walker D, Blair D. Comparisons of ribosomal internal transcribed spacers from two congeneric species of flukes (Platyhelminthes : Trematoda : Digenea)[J]. Mol Biochem Parasitol, 1992, 56(2): 323-327.
doi: 10.1016/0166-6851(92)90181-I |
| [15] | Zhang CL, Qiu YY, Hao S, et al. Development of RPA assay for the detection of Clonorchis sinensis metacercaria[J]. Chin Vet Sci, 2021, 51(4): 441-445. (in Chinese) |
| (张春玲, 邱阳元, 郝朔, 等. 华支睾吸虫囊蚴RPA检测方法的建立[J]. 中国兽医科学, 2021, 51(4): 441-445.) | |
| [16] |
Rahman SMM, Song HB, Jin Y, et al. Application of a loop-mediated isothermal amplification (LAMP) assay targeting cox1 gene for the detection of Clonorchis sinensis in human fecal samples[J]. PLoS Negl Trop Dis, 2017, 11(10): e0005995.
doi: 10.1371/journal.pntd.0005995 |
| [17] | Gao JF, Wang X, Mao RF, et al. Establishment of multiplex PCR for detection of three kinds of Trematode metacercariae in freshwater fish[J]. Chin J Prev Vet Med, 2022, 44(2): 157-161. (in Chinese) |
| (高俊峰, 王鑫, 毛瑞锋, 等. 淡水鱼中3种吸虫囊蚴多重PCR检测方法的建立[J]. 中国预防兽医学报, 2022, 44(2): 157-161.) | |
| [18] |
Lei L, Liao F, Tan L, et al. LAMP coupled CRISPR-Cas12a module for rapid, sensitive and visual detection of porcine circovirus 2[J]. Animals, 2022, 12(18): 2413.
doi: 10.3390/ani12182413 |
| [19] | Zhang SM, Wang G, Cao W, et al. A rare case of co-infection of Clonorchis sinensis, Xenopsylla sinensis and Echinococcus sinensis[J]. Chin J Parasitol Parasit Dis, 2021, 39(4): 561-563. (in Chinese) |
| (张时民, 王庚, 曹玮, 等. 罕见华支睾吸虫、异形吸虫及棘口吸虫合并感染1例[J]. 中国寄生虫学与寄生虫病杂志, 2021, 39(4): 561-563.) | |
| [20] | Mahanta J, Narain K, Srivastava VK. Heterophyid eggs in human stool samples in Assam: first report for India[J]. J Commun Dis, 1995, 27(3): 142-145. |
| [1] | LI Xiaoqin, LAI Yashi, CHEN Yu, LV Jiahui, WEI Shuai, ZHANG Lilin, HE Shanshan, SHI Yunliang, LI Yanwen. Ultrastructural observation on excystment of metacercaria of Clonorchis sinensis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(5): 601-608. |
| [2] | FANG Wen, YANG Jing, WANG Haiying, CHEN Shaorong, LIU Yuhua, LI Tianmei. Excystment conditions and excysted metacercaria morphological observation of Fasciola hepatica [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(2): 198-201. |
| [3] | ZHANG Fengyu, LIU Liu, ZHANG Jing, ZHANG Hao. Prevalence of trematode metacercariae in Pseudorasbora parva and species identification in Qiqihaer area [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(1): 112-116. |
| [4] | HUANG Guang-hua, ZHANG Bo, LAI Hai-tao, Lv Guo-li, LIN Yuan, TANG Wen-qian, WAN Xiao-ling, JIANG Zhi-hua, LIU Jian. Analysis on the prevalence of Clonorchis sinensis infection in Lingshan County, Guangxi from 2016 to 2019 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(5): 673-676. |
| [5] | WANG Ting, YANG Qing-li, LENG Jing, LI Bao-ying, SHEN Ji-qing, DAI Yue. Expression of the high mobility group box 1 homologous protein of Clonorchis sinensis and its effect on nuclear transcription factor-κB activation in mouse macrophages [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(3): 305-308. |
| [6] | CAO Tian-xing, LIU Jun-long, ZHANG Zhi-gang, SHI Kang-yan, SHI Miao, GUAN Gui-quan, LI You-quan, YIN Hong, LUO Jian-xun. Prokaryotic expression of Theileria annulata recombinant TA04380 protein and establishment of ELISA [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(3): 362-368. |
| [7] | JING Wen-wen, CHENG Xun-jia. Application and prospect of multidisciplinary new detection technology in the diagnosis of parasite infections [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(1): 20-27. |
| [8] | TANG Lei, YUAN Shuang, YIN Shi-hui, GE Tao, GE Jing-xue, XING Zhi-feng. Investigation on the prevalence of Clonorchis sinensis infection in human population of Heilongjiang Province, 2015 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(6): 741-745. |
| [9] | CHEN Bao-jian, XIE Han-guo, XIE Xian-liang, JIANG Dian-wei, CHEN Yun-hong, GAO Lan-lin. Investigation of Clonorchis sinensis infection in the national surveillance site of Pucheng County, Fujian Province during 2016—2020 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(5): 716-719. |
| [10] | LI An-mei, HUANG Yu-ting, YAO Dan-cheng, ZHU Ai-ya, LI Yang, SHI Wei-fang, DAI Jia-rui, GENG Yan, SHE Dan-ya, ZHANG Nian-heng. Investigation and analysis on the prevalence of Clonorchis sinensis infection in Guizhou Province in 2015 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(1): 125-128. |
| [11] | YUAN Chang-hong, LAN Ming-xing, ZHU Ting-jun, WANG Mei, CHEN Zhe, HU Li-feng, HUANG Qi, JIANG Wei-sheng. Analysis on the status of human infection in national surveillance sites for clonorchiasis in Xinfeng County during 2016-2019 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(5): 561-564. |
| [12] | HUANG Qi, JIANG Wei-sheng, YAN Yue-kang, CHEN Zhe, XIONG Yan-feng, YANG Yu-hua, WANG Shou-shan. Investigation on the status of Clonorchis sinensis infection in Xinfeng County, Jiangxi Province, China [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(5): 670-673. |
| [13] | Xiao LI, Jiang-chang WANG, Lei GUAN, Qi GU, Ji-ze DONG, Chen-yun WU, Min YAN, Zhao-jun WANG. Advances in the pathogenesis of cholangiocarcinoma caused by Clonorchis sinensis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(2): 250-254. |
| [14] | HU Kun-min, CHEN Shao-hong, AI Lin, ZHENG Bin. Sequence analysis of ribosomal ITS2 gene and mitochondrial CO1 gene of Paragonimus metacercariae from freshwater crabs in Henan, Anhui, Fujian and Zhejiang provinces, China [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(1): 87-94. |
| [15] | Ze-hu SHENG, Shuo ZHANG, Jin-biao YAO, Fan ZHANG, Guan-bei LI, Jia GUO, Hao ZHANG. An investigation of parasitic infections in aquatic products sold in Qiqihar [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2019, 37(5): 616-618. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||