
CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES ›› 2025, Vol. 43 ›› Issue (1): 61-68.doi: 10.12140/j.issn.1000-7423.2025.01.010
• ORIGINAL ARTICLES • Previous Articles Next Articles
DU Jianbo( )(
)( ), SU Yaxin, HUO Lele, WANG Ying, WANG Xu, JIANG Bin, CHEN Yuqing, SHEN Yujuan*(
), SU Yaxin, HUO Lele, WANG Ying, WANG Xu, JIANG Bin, CHEN Yuqing, SHEN Yujuan*( )(
)( )
)
Received:2024-12-05
Revised:2025-01-07
Online:2025-02-28
Published:2025-03-26
Contact:
E-mail: Supported by:CLC Number:
DU Jianbo, SU Yaxin, HUO Lele, WANG Ying, WANG Xu, JIANG Bin, CHEN Yuqing, SHEN Yujuan. Establishment and application of a dual-probe fluorescent recombinase polymerase amplification assay for detection of Echinococcus granulosus[J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2025, 43(1): 61-68.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.jsczz.cn/EN/10.12140/j.issn.1000-7423.2025.01.010
Table 1
Sequence of primers and fluorescent probes
| 引物及探针名称 Name of primers and probes | 序列(5′→3′) Sequence (5′→3′) |
|---|---|
| co1-F | TGTTTGGGTTCTATGGGTTGTTGTTTGCTATGT |
| co1-R | AACACACAAGCAGACAAAACTATACCCGTAACTC |
| nd5-F | GGCGGTGATTATGTTGGGTTTATGTTGTTGAAGT |
| nd5-R | AACCAAACACCAGCAGCAACCAAAGTAGAAG |
| HEX-P | AGAGTGCTGGTTATCCTTTTATTAGGTGG(iHEXdT)T(idsp)(iBHQ1dT)TGGAGGCGATGCGGGC-C3 Spacer |
| FAM-P | GGTAGCAGGGTTTGGGGTCATCATATGT(i6FAMdT)T(idsp)C(iBHQ1dT)GTTGGGTTGGATGTGAA-C3 Spacer |
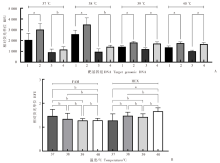
Fig. 2
Effect of temperature on the dual-probe fluorescent RPA assay A: Effect of temperature on amplification of E. granulosus genomic DNA with the dual-probe fluorescent RPA assay; B: Ratio of the amplification fluorescence signal of the E. granulosus target gene to that of the blank control at different temperatures. 1: Blank control (FAM); 2: E. granulosus (FAM);3: Blank control (HEX); 4: E. granulosus (HEX). a: P < 0.01; b: P > 0.01.
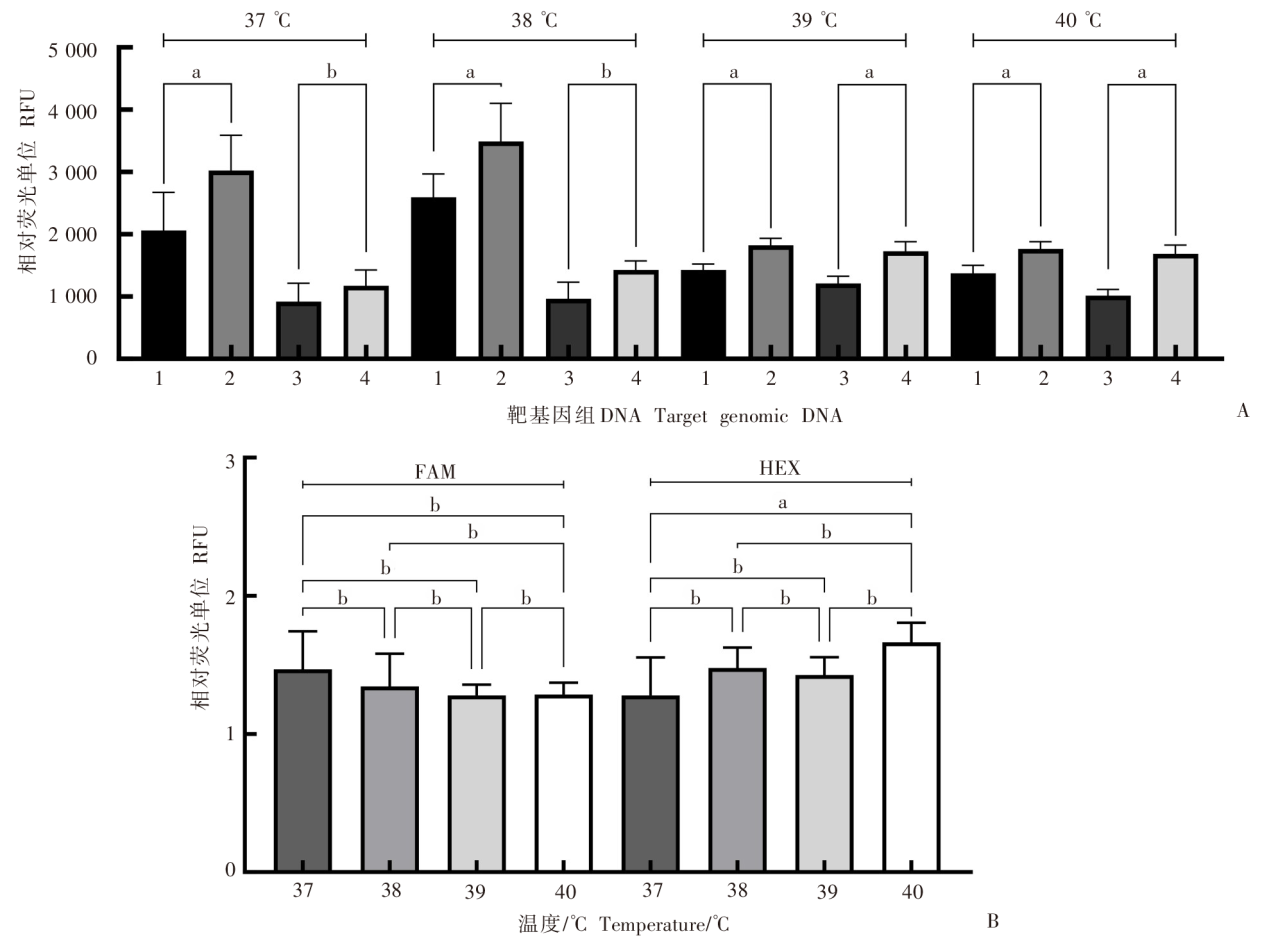

Fig. 3
Sensitivity of the dual-probe fluorescent RPA assay for detecting recombinant plasmids containing co1、nd5 A: Amplification signal intensity of different concentrations of plasmids pUC-SP-co1 with FAM fluorescence; B: Amplification signal intensity of different concentrations of plasmids pUC57-nd5 with FAM fluorescence. a: P < 0.01; b: P > 0.01.
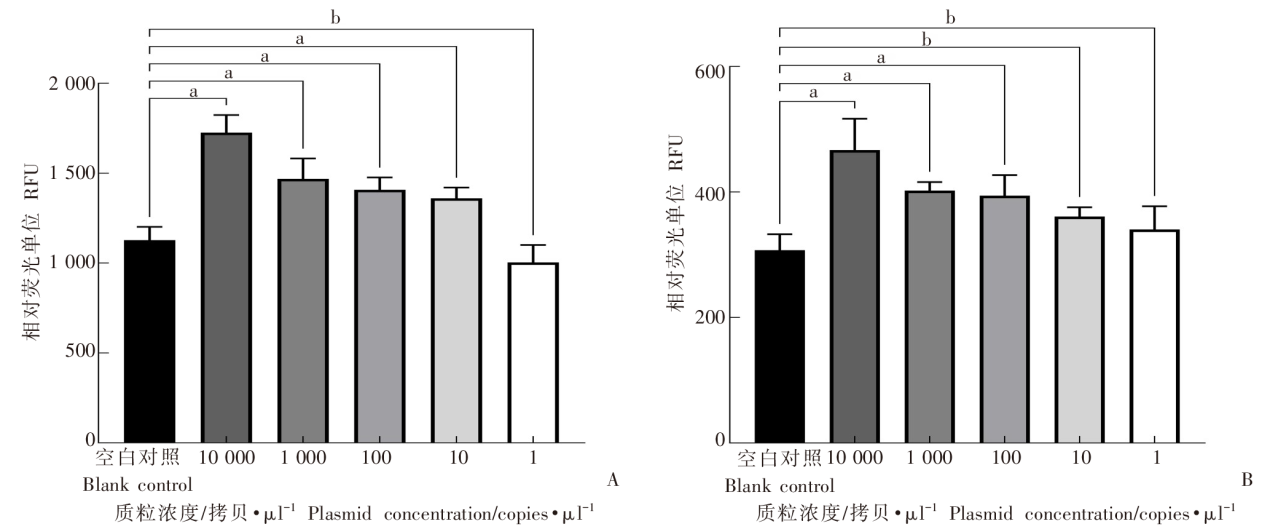
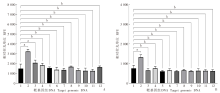
Fig. 4
Specificity of the dual-probe fluorescent RPA assay for detecting E. granulosus a: P < 0.01; b: P > 0.01. A: Relative FAM fluorescence intensity for amplification of genomic DNA from different parasites with the dual-probe fluorescent RPA assay; B: Relative HEX fluorescence intensity for amplification of genomic DNA from different parasites with the dual-probe fluorescent RPA assay. 1: Blank control; 2: E. granulosus; 3: E. multilocularis; 4: T. solium; 5: S. japonicum; 6: N. americanus; 7: N. braziliensis; 8: C. sinensis; 9: G. lamblia; 10: C. parvum; 11: T. gondii; 12: B. microti.
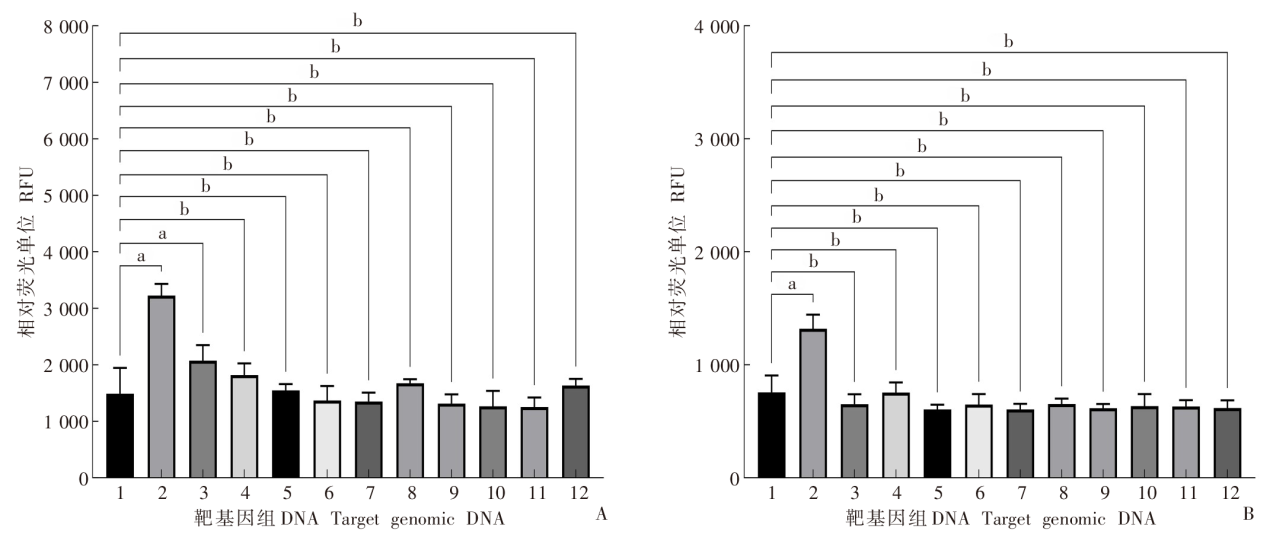
Table 2
Comparison of the detection efficiency between the dual-probe fluorescent RPA assay and PCR assay
| 样品 Sample | 双探针荧光RPA方法 Dual-probe fluorescent RPA assay | PCR | |||
|---|---|---|---|---|---|
| FAM/HEX | FAM | 阴性 Negative | 阳性 Positive | ||
| 细粒棘球绦虫模拟样品 E. granulosus simulation sample | 8 | 0 | 0 | 8 | |
| 多房棘球绦虫模拟样品 E. multilocularis simulation sample | 0 | 7 | 0 | 7 | |
| 细粒棘球绦虫和多房棘球绦虫混合模拟样品 E. granulosus + E. multilocularis simulation sample | 8 | 0 | 0 | 8 | |
| 犬粪样 Dog fecal sample | 5 | 0 | 0 | 5 | |
| 合计 Total | 21 | 7 | 0 | 28 | |
| [1] | Wen H, Vuitton L, Tuxun T, et al. Echinococcosis: Advances in the 21st century[J]. Clin Microbiol Rev, 2019, 32(2): e00075-18. |
| [2] |
蒉嫣, 薛垂召, 王旭, 等. 2022年全国棘球蚴病防治工作进展[J]. 中国寄生虫学与寄生虫病杂志, 2024, 42(1): 8-16.
doi: 10.12140/j.issn.1000-7423.2024.01.002 |
| Kui Y, Xue CZ, Wang X, et al. Progress of echinococcosis control in China, 2022[J]. Chin J Parasitol Parasit Dis, 2024, 42(1): 8-16. (in Chinese) | |
| [3] | Wang ZH, Wang XM, Liu XQ. Echinococcosis in China, a review of the epidemiology of Echinococcus spp.[J]. Ecohealth, 2008, 5(2): 115-126. |
| [4] | Vola A, Manciulli T, De Silvestri A, et al. Diagnostic performances of commercial ELISA, indirect hemagglutination, and western blot in differentiation of hepatic echinococcal and non-echinococcal lesions: A retrospective analysis of data from a single referral centre[J]. Am J Trop Med Hyg, 2019, 101(6): 1345-1349. |
| [5] | Lobato IM, O’Sullivan CK. Recombinase polymerase amplification: Basics, applications and recent advances[J]. Trends Analyt Chem, 2018, 98: 19-35. |
| [6] | Liu XQ, Yan QY, Huang JF, et al. Influence of design probe and sequence mismatches on the efficiency of fluorescent RPA[J]. World J Microbiol Biotechnol, 2019, 35(6): 95. |
| [7] |
Nakao M, McManus DP, Schantz PM, et al. A molecular phylogeny of the genus Echinococcus inferred from complete mitochondrial genomes[J]. Parasitology, 2007, 134(Pt 5): 713-722.
doi: 10.1017/S0031182006001934 pmid: 17156584 |
| [8] | Alvarez Rojas CA, Romig T, Lightowlers MW. Echinococcus granulosus sensulato genotypes infecting humans: Review of current knowledge[J]. Int J Parasitol, 2014, 44(1): 9-18. |
| [9] |
Khan SN, Ali R, Khan S, et al. Cystic echinococcosis: An emerging zoonosis in southern regions of Khyber Pakhtunkhwa, Pakistan[J]. BMC Vet Res, 2021, 17(1): 139.
doi: 10.1186/s12917-021-02830-z pmid: 33794898 |
| [10] |
Moro P, Schantz PM. Echinococcosis: A review[J]. Int J Infect Dis, 2009, 13(2): 125-133.
doi: 10.1016/j.ijid.2008.03.037 pmid: 18938096 |
| [11] | Shang JY, Zhang GJ, Liao S, et al. A multiplex PCR for differential detection of Echinococcus granulosus sensustricto, Echinococcus multilocularis and Echinococcus canadensis in China[J]. Infect Dis Poverty, 2019, 8: 68. |
| [12] | Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA[J]. Nucleic Acids Res, 2000, 28(12): E63. |
| [13] | 张艳艳, 叶倩, 王正荣, 等. 基于cox2基因的细粒棘球绦虫环介导等温扩增检测方法的初步建立[J]. 中国寄生虫学与寄生虫病杂志, 2017, 35(2): 169-172. |
| Zhang YY, Ye Q, Wang ZR, et al. Preliminary exploration of loop-mediated isothermal amplification based on cox2 gene of Echinococcus granulosus[J]. Chin J Parasitol Parasit Dis, 2017, 35(2): 169-172. (in Chinese) | |
| [14] | 吕蓓, 程海荣, 严庆丰, 等. 用重组酶介导扩增技术快速扩增核酸[J]. 中国科学: 生命科学, 2010, 40(10): 983-988. |
| Lü B, Cheng HR, Yan QF, et al. Recombinase-aid amplification: A novel technology of in vitro rapid nucleic acid amplification[J]. Sci Sin Vitae, 2010, 40(10): 983-988. (in Chinese) | |
| [15] | 李婷, 杨坤. 等温扩增技术在寄生虫及其他病原体检测中的应用[J]. 中国血吸虫病防治杂志, 2018, 30(2): 232-236. |
| Li T, Yang K. Application of isothermal amplification technology for pathogen detection in parasitic and other diseases[J]. Chin J Schisto Control, 2018, 30(2): 232-236. (in Chinese) | |
| [16] |
周鸿让, 茅光耀, 王晓玲, 等. 重组酶介导的多重核酸等温扩增法鉴别细粒棘球绦虫和多房棘球绦虫技术的建立与应用[J]. 中国寄生虫学与寄生虫病杂志, 2020, 38(3): 310-316.
doi: 10.12140/j.issn.1000-7423.2020.03.09 |
| Zhou HR, Mao GY, Wang XL, et al. Establishment and application of a multiplex recombinase-aided isothermal amplification technique for identifying Echinococcus granulosus and Echinococcus multilocularis[J]. Chin J Parasitol Parasit Dis, 2020, 38(3): 310-316. (in Chinese) | |
| [17] | Teoh BT, Sam SS, Tan KK, et al. Early detection of dengue virus by use of reverse transcription-recombinase polymerase amplification[J]. J Clin Microbiol, 2015, 53(3): 830-837. |
| [18] |
梁家瑞, 徐斌, 胡薇, 等. 基于荧光重组酶聚合酶扩增技术快速检测美洲钩虫方法研究[J]. 中国热带医学, 2023, 23(7): 681-685.
doi: 10.13604/j.cnki.46-1064/r.2023.07.01 |
| Liang JR, Xu B, Hu W, et al. Development and preliminary evaluation of a fluorescence RPA assay for the rapid detection of Necator americanus[J]. China Trop Med, 2023, 23(7): 681-685. (in Chinese) | |
| [19] | Tian LB, Shi Y, Yang Y, et al. Rapid on-site detection of echinococcosis and schistosomiasis based on RPA[J]. Mem Inst Oswaldo Cruz, 2024, 119: e230244. |
| [20] | 苏书晓, 赵帅阳, 刘军龙, 等. 环形泰勒虫SHERLOCK-LF检测方法的建立与初步应用[J]. 中国兽医科学, 2024, 54(9): 1182-1187. |
| Su SX, Zhao SY, Liu JL, et al. Establishment and preliminary application of SHERLOCK-LF detection method for Theileria annulata[J]. Chin Vet Sci, 2024, 54(9): 1182-1187. (in Chinese) | |
| [21] | 高璟瑜, 李秋阳, 姚瑶, 等. 恶性疟原虫LF-RPA检测方法研究[J]. 寄生虫与医学昆虫学报, 2023, 30(4): 193-196, 209. |
| Gao JY, Li QY, Yao Y, et al. Study on LF-RPA detection method of Plasmodium falciparum[J]. Acta Parasitol Med Entomol Sin, 2023, 30(4): 193-196, 209. (in Chinese) | |
| [22] |
王盛琳, 邓王平, 李银龙, 等. 重组酶聚合酶扩增技术快速检测日本血吸虫核酸方法的建立[J]. 中国寄生虫学与寄生虫病杂志, 2020, 38(3): 293-298.
doi: 10.12140/j.issn.1000-7423.2020.03.006 |
| Wang SL, Deng WP, Li YL, et al. Establishment of recombinase polymerase amplification technique for rapid detection of Schistosoma japonicum nucleic acid[J]. Chin J Parasitol Parasit Dis, 2020, 38(3): 293-298. (in Chinese) | |
| [23] |
王丽萍, 吕超, 秦志强, 等. 基于重组酶聚合酶扩增的曼氏血吸虫核酸可视化检测技术的建立及初步评价[J]. 中国寄生虫学与寄生虫病杂志, 2022, 40(3): 337-343.
doi: 10.12140/j.issn.1000-7423.2022.03.009 |
| Wang LP, Lv C, Qin ZQ, et al. Establishment and preliminary evaluation of a visualized detection technique for Schistosoma mansoni nucleic acid based on recombinase polymerase amplification[J]. Chin J Parasitol Parasit Dis, 2022, 40(3): 337-343. (in Chinese) |
| [1] | YAN Shuning, YANG Hanyin, CAI Yuchun, XU Bin, YU Chenghang, MO Ziran, LU Yan, YANG Shuo, XIN Yi, ZHENG Bin. Establishment and assessment of nucleic acid detection method for Ancylostoma duodenale based on RPA-CRISPR/Cas12a technology [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(6): 748-755. |
| [2] | XU Guolei, FENG Yanye, HU Wei. Establishment and application evaluation of a rapid visualization detection method for Schistosoma japonicum specific nucleic acid fragments based on RPA-CRISPR/Cas12a technology [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(5): 608-614. |
| [3] | WANG Xu, WANG Ying, LIU Baixue, ZHANG Kaige, DENG Xueying, SHEN Yujuan, WANG Zhenghuan, CAO Jianping, HAN Shuai. A brief cognitive and historical overview of Echinococcus and echinococcosis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(3): 372-383. |
| [4] | JIANG Xiao-feng, SHEN Yu-juan. Research progress on liver fibrosis caused by Echinococcus infection [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(5): 656-660. |
| [5] | ZUO Qing-qiu, ZHENG Jia-xin, WANG Gang, WANG Xu, WANG Xiao-ming, WANG Zheng-huan. The transimission ecology of the echinococcosis on the Tibetan Plateau [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(4): 524-535. |
| [6] | XILIZATI Kulaixi, WANG Chun-sheng, WANG Jia-ling, LI Meng, FANG Zhi-yuan, WANG Si-jia, ZHOU Jing-ru, XIANYIDAN Abulajiang, WU Er-ge-li, YE Jian-rong. Bioinformatics analysis of the sensitization mechanisms and molecular targets of Echinococcus granulosus [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(3): 319-323. |
| [7] | XU Kai, HUANG Lu-lu, WU Chuan-ling, LI Wen-deng, YIN Feng-jiao, WANG Zhi-xin, FAN Hai-ning, WANG Hai-jiu. Research progress on the experimental treatment of Echinococcus infection by using Chinese traditional medicine [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(5): 710-715. |
| [8] | WANG Qi, WANG Qian, CHEN Shun-de, HE Wei, YU Wen-jie, YANG Liu, ZHANG Guang-jia, LIAO Sha, LI Rui-rui, HUANG Yan, YAO Ren-xin, LIU Yang, ZHONG Bo. Observation on the behavioral activities of the wild animals as intermediate hosts for Echinococcus spp. by infrared-triggered cameras in Shiqu County of Sichuan Province [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(2): 178-184. |
| [9] | CUI Xiao-yu, GUAN Ya-yi, WU Wei-ping. The prevalence and impacting factors of Echinococcus infection in canine in Qinghai-Tibet plateau [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(4): 482-489. |
| [10] | DENG Wang-ping, HONG Qing-hua, XU Bin, WANG Sheng-lin, WANG Li-ping, XU Jing, HU Wei, ZHOU Xiao-nong. Development and preliminary evaluation of a rapid visualization detection method for circulating nucleic acids of Schistosoma japonicum based on RPA-LFD [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(3): 286-292. |
| [11] | WANG Sheng-lin, DENG Wang-ping, LI Yin-long, WANG Li-ping, ZHANG Li-juan, LV Shan, XU Jing. Establishment of recombinase polymerase amplification technique for rapid detection of Schistosoma japonicum nucleic acid [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(3): 293-298. |
| [12] | Wang-ping DENG, Bin XU, Qing-hua HONG, Sheng-lin WANG, Chao LV, Yin-long LI, Shi-ping SONG, Jun-hu CHEN, Jing XU, Shi-zhu LI, Wei HU, Xiao-nong ZHOU. Establishment of the detection method for Schistosoma japonicum by recombinase polymerase amplification combined with electrochemical DNA biosensor [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(2): 168-174. |
| [13] | Qi WANG, Shun-de CHEN, Qian WANG, Wei HE, Wen-jie YU, Guang-jia ZHANG, Fan CHEN, Liu YANG, Sha LIAO, Rui-rui Li, Yan HUANG, Ren-xin YAO, Bo ZHONG. Activities of host animals of Echinococcus spp observed by infrared triggered cameras in Shiqu, Sichuan [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2019, 37(5): 622-625. |
| [14] | Hong-rang ZHOU, Mu-xin CHEN, Ying WANG, Qing YU, Ning XIAO. Research progress on detection technology of Echinococcus and echinococcosis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2019, 37(3): 346-352. |
| [15] | Ling-ling WANG, Hai-mei MA, Jian-bing DING. Research advances on TGF-β/Smad and MAPK signal pathways in Echinococcus [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2018, 36(5): 516-519. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||