
CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES ›› 2024, Vol. 42 ›› Issue (4): 475-480.doi: 10.12140/j.issn.1000-7423.2024.04.008
• ORIGINAL ARTICLES • Previous Articles Next Articles
WANG Jufeng( ), XUE Shijie, LIU Jiangyong, LAI Dehua*(
), XUE Shijie, LIU Jiangyong, LAI Dehua*( ), LUN Zhaorong
), LUN Zhaorong
Received:2024-03-29
Revised:2024-05-17
Online:2024-08-30
Published:2024-08-20
Contact:
E-mail: Supported by:CLC Number:
WANG Jufeng, XUE Shijie, LIU Jiangyong, LAI Dehua, LUN Zhaorong. Infection and identification of Clonorchis sinensis metacercariae in Pseudorasbora parva in multiple regions of China[J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(4): 475-480.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.jsczz.cn/EN/10.12140/j.issn.1000-7423.2024.04.008
Table 1
Clonorchis sinensis metacercariae infection in Pseudorasbora parva from various regions of China
| 采样点 Sampling site | 采样时间 Sampling time | 检查数量/尾 No. examined | 阳性数/尾(阳性率/%) No. positive (Positive rate/%) |
|---|---|---|---|
| 华东地区 East China | |||
| 安徽阜阳 Fuyang, Anhui | 2023.04.28 | 12 | 12(12/12) |
| 安徽阜阳 Fuyang, Anhui | 2024.01.20 | 20 | 12(60) |
| 福建宁德 Ningde, Fujian | 2023.05.13 | 19 | 5(5/19) |
| 浙江衢州 Quzhou, Zhejiang | 2023.05.13 | 40 | 0(0) |
| 山东烟台 Yantai, Shandong | 2023.04.26 | 51 | 49(96.1) |
| 山东烟台 Yantai, Shandong | 2023.09.24 | 23 | 23(100) |
| 山东烟台 Yantai, Shandong | 2023.11.27 | 50 | 50(100) |
| 山东青岛 Qingdao, Shandong | 2022.11.07 | 23 | 1(4.3) |
| 上海 Shanghai | 2023.11.20 | 40 | 0(0) |
| 华南地区 South China | |||
| 广东汕头 Shantou, Guangdong | 2022.11.04 | 50 | 0(0) |
| 广东汕头 Shantou, Guangdong | 2023.05.29 | 32 | 0(0) |
| 广西横州 Hengzhou, Guangxi | 2022.08.01 | 6 | 6(6/6) |
| 华中地区 Central China | |||
| 河南濮阳 Puyang, Henan | 2022.10.15 | 22 | 22(100) |
| 湖北武汉 Wuhan, Hubei | 2023.01.19 | 30 | 0(0) |
| 湖北武汉 Wuhan, Hubei | 2023.04.25 | 36 | 0(0) |
| 西北地区 Northwest China | |||
| 陕西汉中 Hanzhong, Shaanxi | 2022.10.25 | 60 | 60(100) |
| 陕西汉中 Hanzhong, Shaanxi | 2023.04.28 | 50 | 33(66) |
| 陕西汉中 Hanzhong, Shaanxi | 2023.06.02 | 32 | 1(3.1) |
| 陕西汉中 Hanzhong, Shaanxi | 2023.09.18 | 35 | 1(2.9) |
| 陕西汉中 Hanzhong, Shaanxi | 2023.11.20 | 16 | 0(0) |
| 西南地区 Southwest China | |||
| 四川成都 Chengdu, Sichuan | 2023.05.13 | 30 | 2(6.7) |
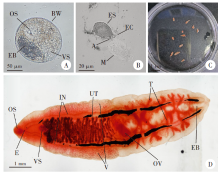
Fig. 1
Morphology of C. sinensis in different life cycle stages A: Metacercaria (× 40); B: Egg, miracidium (× 40); C: Adults collected from a rat bile duct; D: Magnified adult. OS: Oral sucker; VS: Ventral sucker; EB: Excretory bladder; BW: Bladder wall; ES: Egg shell; EC: Egg cap; A: Acromion; M: Miracidium; E: Esophagus; IN: Intestinalis; UT: Uterus; T: Testicles; V: Vitellarium; OV: Ovary.
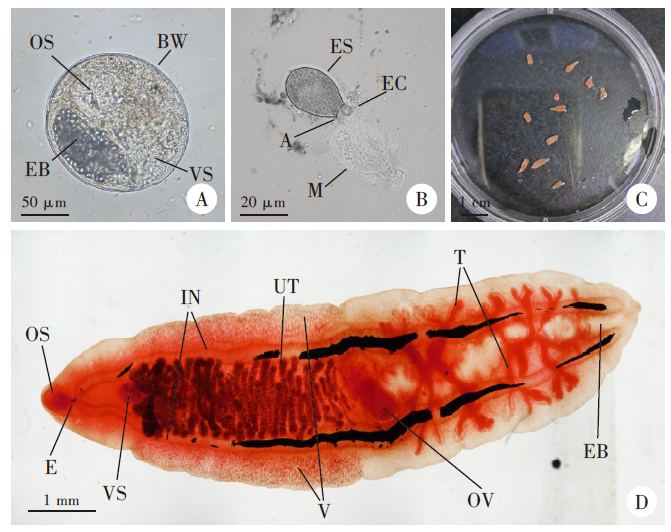
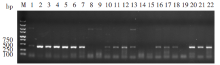
Fig. 2
16S rDNA amplificon from some metacercariae by PCR M: DNA marker; 1-7, 20-22: Samples collected from Yantai, Shandong; 8-9: Samples collected from Fuyang, Anhui; 10-11: Samples collected from Hanzhong, Shaanxi; 12-15: Samples collected from Qingdao, Shandong; 16-19: Samples collected from Puyang, He’nan.
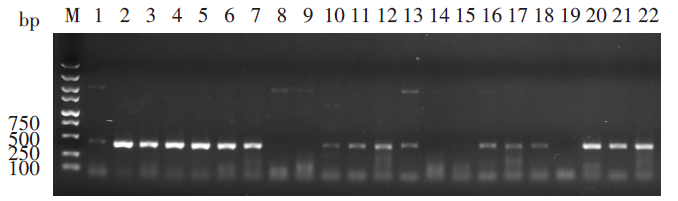

Fig. 3
Phylogenetic tree based on 16S rDNA sequence of Opisthorchiidae using maximum likelihood method Bootstrap values are shown as circles at the nodes, with larger circles indicating higher support. The branch lengths are proportional to the number of nucleotide substitutions per site. ▲Metacercariae samples obtainedinthis study.

| [1] | 2016 DALYs and HALE Collaborators. Global, regional, and national disability-adjusted life-years (DALYs) for 333 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016[J]. Lancet, 2017, 390(10100): 1260-1344. |
| [2] | WHO. Foodborne trematode infections[EB/OL]. (2021-05-21)[2024-03-20]. https://www.who.int/news-room/fact-sheets/detail/foodborne-trematode-infections. |
| [3] | Na BK, Pak JH, Hong SJ. Clonorchis sinensis and clonorchiasis[J]. Acta Trop, 2020, 203: 105309. |
| [4] | Lun ZR, Gasser RB, Lai DH, et al. Clonorchiasis: a key foodborne zoonosis in China[J]. Lancet Infect Dis, 2005, 5(1): 31-41. |
| [5] | Chen YD, Zhou CH, Xu LQ. Analysis of the results of two nationwide surveys on Clonorchis sinensis infection in China[J]. Biomed Environ Sci, 2012, 25(2): 163-166. |
| [6] | Chen YD, Zhou CH, Zhu HH, et al. National survey on the current status of important human parasitic diseases in China in 2015[J]. Chin J Parasitol Parasit Dis, 2020, 38(1): 5-16. (in Chinese) |
|
(陈颖丹, 周长海, 朱慧慧, 等. 2015年全国人体重点寄生虫病现状调查分析[J]. 中国寄生虫学与寄生虫病杂志, 2020, 38(1): 5-16.)
doi: 10.12140/j.issn.1000-7423.2020.01.002 |
|
| [7] | Qian MB, Chen J, Bergquist R, et al. Neglected tropical diseases in the People’s Republic of China: progress towards elimination[J]. Infect Dis Poverty, 2019, 8(1): 86. |
| [8] | Lai DH, Hong XK, Su BX, et al. Current status of Clonorchis sinensis and clonorchiasis in China[J]. Trans R Soc Trop Med Hyg, 2016, 110(1): 21-27. |
| [9] | Tang ZL, Huang Y, Yu XB. Current status and perspectives of Clonorchis sinensis and clonorchiasis: epidemiology, pathogenesis, omics, prevention and control[J]. Infect Dis Poverty, 2016, 5(1): 71. |
| [10] | Sohn WM. Infection characteristics of Clonorchis sinensis metacercariae in fish from Republic of Korea[J]. Korean J Parasitol, 2022, 60(2): 79-96. |
| [11] | Chen DX, Chen JY, Huang J, et al. Epidemiological investigation of Clonorchis sinensis infection in freshwater fishes in the Pearl River Delta[J]. Parasitol Res, 2010, 107(4): 835-839. |
| [12] | Xie YH, Kanankege KST, Jiang ZH, et al. Epidemiological characterization of Clonorchis sinensis infection in humans and freshwater fish in Guangxi, China[J]. BMC Infect Dis, 2022, 22(1): 263. |
| [13] | Qiu JH, Zhang Y, Zhang XX, et al. Metacercaria infection status of fishborne zoonotic trematodes, except for Clonorchis sinensis in fish from the Heilongjiang Province, China[J]. Foodborne Pathog Dis, 2017, 14(8): 440-446. |
| [14] | Sohn WM. Fish-borne zoonotic trematode metacercariae in the Republic of Korea[J]. Korean J Parasitol, 2009, 47(Suppl): S103-S113. |
| [15] | Sohn WM, Eom KS, Min DY, et al. Fishborne trematode metacercariae in freshwater fish from Guangxi Zhuang Autonomous Region, China[J]. Korean J Parasitol, 2009, 47(3): 249-257. |
| [16] | Lai DH, Wang QP, Chen W, et al. Molecular genetic profiles among individual Clonorchis sinensis adults collected from cats in two geographic regions of China revealed by RAPD and MGE-PCR methods[J]. Acta Trop, 2008, 107(2): 213-216. |
| [17] |
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Mol Biol Evol, 2016, 33(7): 1870-1874.
doi: 10.1093/molbev/msw054 pmid: 27004904 |
| [18] | Price MN, Dehal PS, Arkin AP. FastTree 2: approximately maximum-likelihood trees for large alignments[J]. PLoS One, 2010, 5(3): e9490. |
| [19] | Sun T, Bao GL, Hu CD, et al. Detection ofeggs in livestock manure[J]. Chin J Anim Husb Vet Med, 2016(6): 35. (in Chinese) |
| (孙涛, 包广玲, 胡成栋, 等. 畜禽粪便内虫卵检查法[J]. 畜牧兽医科技信息, 2016(6): 35.) | |
| [20] | Xie WT, Shuai ML, Wang SS, et al. Exploration on the technology of counter staining of fluke[J]. Chin J Parasitol Parasit Dis, 2016, 34(4): 323-325. (in Chinese) |
| (谢文婷, 帅木兰, 王少圣, 等. 两种吸虫复染制片技术的探究[J]. 中国寄生虫学与寄生虫病杂志, 2016, 34(4): 323-325.) | |
| [21] | Qiao T, Ma RH, Luo XB, et al. Cholecystolithiasis is associated with Clonorchis sinensis infection[J]. PLoS One, 2012, 7(8): e42471. |
| [22] | Ding YL, Xia P, Zhang XF, et al. Etiological investigation on food-derived parasites in farm produce market of Jingzhou city[J]. J Anhui Agric Sci, 2010, 38(24): 13124-13125. (in Chinese) |
| (丁义玲, 夏鹏, 张晓方, 等. 荆州市农贸市场食源性寄生虫病原学调查[J]. 安徽农业科学, 2010, 38(24): 13124-13125.) | |
| [23] | Zhu H, Sun CS, Wang QZ, et al. Survey of Clonorchis sinensis infection in freshwater fish in some areas in Anhui Province[J]. J Trop Dis Parasitol, 2022, 20(6): 325-329. (in Chinese) |
| (朱海, 孙成松, 汪奇志, 等. 安徽省部分地区淡水鱼华支睾吸虫感染状况调查分析[J]. 热带病与寄生虫学, 2022, 20(6): 325-329.) | |
| [24] | Geng YZ, Yu M, Zhang MY, et al. Investigation of trematode metacercariae infection in freshwater fish in Liaoning Province[J]. J Trop Med, 2022, 22(6): 868-871. (in Chinese) |
| (耿英芝, 于淼, 张铭琰, 等. 辽宁省淡水鱼吸虫囊蚴感染调查[J]. 热带医学杂志, 2022, 22(6): 868-871.) | |
| [25] | Zhu TJ, Li HM, Wang JJ, et al. Infection status of freshwater fishes with metacercariae of fish born trematode in Heng County Guangxi[J]. Int J Med Parasit Dis, 2012, 39(3): 167-170. (in Chinese) |
| (诸廷俊, 李红梅, 王聚君, 等. 广西横县淡水鱼吸虫囊蚴感染情况调查[J]. 国际医学寄生虫病杂志, 2012, 39(3): 167-170.) | |
| [26] | Sheng ZH, Zhang S, Yao JB, et al. An investigation of parasitic infections in aquatic products sold in Qiqihar[J]. Chin J Parasitol Parasit Dis, 2019, 37(5): 616-618. (in Chinese) |
|
(盛泽虎, 张硕, 姚金彪, 等. 齐齐哈尔地区市售水产品中寄生虫感染情况调查[J]. 中国寄生虫学与寄生虫病杂志, 2019, 37(5): 616-618.)
doi: 10.12140/j.issn.1000-7423.2019.05.021 |
|
| [27] | Wang M, Luo L, Chen XQ, et al. Investigation on sanitation of freshwater aquaculture environments and Clonorchis sinensis intermediate host infection in a city of Pearl River Delta region[J]. Chin J Schisto Control, 2017, 29(6): 716-719. (in Chinese) |
| (王曼, 罗乐, 陈雪琴, 等. 珠三角某市淡水鱼养殖环境及华支睾吸虫感染情况调查[J]. 中国血吸虫病防治杂志, 2017, 29(6): 716-719.) | |
| [28] | Liu JX, Sun YH, Zhang H, et al. Prevalence of metacercariae of Clonorchis sinensis in wild freshwater fishes from Nenjiang River around Qiqihaer City[J]. Chin J Parasitol Parasit Dis, 2014, 32(4): 292-294. (in Chinese) |
| (刘继鑫, 孙艳宏, 张浩, 等. 嫩江流域齐齐哈尔段野生淡水鱼华支睾吸虫囊蚴感染现状[J]. 中国寄生虫学与寄生虫病杂志, 2014, 32(4): 292-294.) | |
| [29] | Zhang FY, Liu L, Zhang J, et al. Prevalence of trematode metacercariae in Pseudorasbora parva and species identification in Qiqihaer Area[J]. Chin J Parasitol Parasit Dis, 2023, 41(1): 112-116. (in Chinese) |
|
(张凤玉, 刘柳, 张静, 等. 齐齐哈尔地区麦穗鱼吸虫囊蚴感染情况及其种类鉴定[J]. 中国寄生虫学与寄生虫病杂志, 2023, 41(1): 112-116.)
doi: 10.12140/j.issn.1000-7423.2023.01.018 |
|
| [30] |
Binkienė R, Miliūtė A, Stunžėnas V. Molecular data confirm the taxonomic position of Hymenolepis erinacei (Cyclophyllidea ∶ Hymenolepididae) and host switching, with notes on cestodes of Palaearctic hedgehogs (Erinaceidae)[J]. J Helminthol, 2019, 93(2): 195-202.
doi: 10.1017/S0022149X18000056 pmid: 29386083 |
| [31] | Gong SH, Ding YF, Wang Y, et al. Advances in DNA barcoding of toxic marine organisms[J]. Int J Mol Sci, 2018, 19(10): 2931. |
| [1] | CHAI Yingzhi, CHEN Hualiang, YU Kegen, ZHANG Xuan, WANG Xiaoxiao, ZHANG Jiaqi, XU Wenjie, RUAN Wei. Analysis on surveillance results of clonorchiasis in Zhejiang Province from 2013 to 2022 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(4): 469-474. |
| [2] | LI Jianke, ZHANG Jing, LIU Liu, LIU Qianhao, ZHANG Hao. Molecular identification of four species of trematode larvae in freshwater snails from Qiqihar area [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(3): 360-366. |
| [3] | LIU Liu, ZHANG Jing, LI Jianke, ZHANG Hao, ZHANG Fengyu. Distribution of the neurotransmitter 5-hydroxytryptamine in Clonorchis sinensis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(1): 78-82. |
| [4] | ZHAO Lei, LI Jia, MO Gang, LI Chun, HUANG Guoyang, PENG Xiaohong. Effect of Clonorchis sinensis infection on hepatic fibrosis and immune regulation in mice [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(6): 760-765. |
| [5] | LI Xiaoqin, LAI Yashi, CHEN Yu, LV Jiahui, WEI Shuai, ZHANG Lilin, HE Shanshan, SHI Yunliang, LI Yanwen. Ultrastructural observation on excystment of metacercaria of Clonorchis sinensis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(5): 601-608. |
| [6] | XU Yin, LIU Ting, XU Hui, ZENG Xiaojun, LAN Weiming, GONG Zhihong, DAI Kunjiao, QIU Tingting, HAO Xian, XIE Shuying. Establishment and application of PCR-CRISPR/Cas12a-based detection method for Clonorchis sinensis metacercaria [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(4): 421-426. |
| [7] | FANG Wen, YANG Jing, WANG Haiying, CHEN Shaorong, LIU Yuhua, LI Tianmei. Excystment conditions and excysted metacercaria morphological observation of Fasciola hepatica [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(2): 198-201. |
| [8] | ZHANG Fengyu, LIU Liu, ZHANG Jing, ZHANG Hao. Prevalence of trematode metacercariae in Pseudorasbora parva and species identification in Qiqihaer area [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(1): 112-116. |
| [9] | HUANG Guang-hua, ZHANG Bo, LAI Hai-tao, Lv Guo-li, LIN Yuan, TANG Wen-qian, WAN Xiao-ling, JIANG Zhi-hua, LIU Jian. Analysis on the prevalence of Clonorchis sinensis infection in Lingshan County, Guangxi from 2016 to 2019 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(5): 673-676. |
| [10] | WANG Ting, YANG Qing-li, LENG Jing, LI Bao-ying, SHEN Ji-qing, DAI Yue. Expression of the high mobility group box 1 homologous protein of Clonorchis sinensis and its effect on nuclear transcription factor-κB activation in mouse macrophages [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(3): 305-308. |
| [11] | TANG Lei, YUAN Shuang, YIN Shi-hui, GE Tao, GE Jing-xue, XING Zhi-feng. Investigation on the prevalence of Clonorchis sinensis infection in human population of Heilongjiang Province, 2015 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(6): 741-745. |
| [12] | CHEN Bao-jian, XIE Han-guo, XIE Xian-liang, JIANG Dian-wei, CHEN Yun-hong, GAO Lan-lin. Investigation of Clonorchis sinensis infection in the national surveillance site of Pucheng County, Fujian Province during 2016—2020 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(5): 716-719. |
| [13] | LI An-mei, HUANG Yu-ting, YAO Dan-cheng, ZHU Ai-ya, LI Yang, SHI Wei-fang, DAI Jia-rui, GENG Yan, SHE Dan-ya, ZHANG Nian-heng. Investigation and analysis on the prevalence of Clonorchis sinensis infection in Guizhou Province in 2015 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(1): 125-128. |
| [14] | YUAN Chang-hong, LAN Ming-xing, ZHU Ting-jun, WANG Mei, CHEN Zhe, HU Li-feng, HUANG Qi, JIANG Wei-sheng. Analysis on the status of human infection in national surveillance sites for clonorchiasis in Xinfeng County during 2016-2019 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(5): 561-564. |
| [15] | HUANG Qi, JIANG Wei-sheng, YAN Yue-kang, CHEN Zhe, XIONG Yan-feng, YANG Yu-hua, WANG Shou-shan. Investigation on the status of Clonorchis sinensis infection in Xinfeng County, Jiangxi Province, China [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(5): 670-673. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||