
CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES ›› 2017, Vol. 35 ›› Issue (3): 280-287.
• Original Articles • Previous Articles Next Articles
Yu-yan GUO1, Lei LUO2, Zhang-yao SONG1, Xue-li ZHENG1,*( )
)
Received:2017-02-24
Online:2017-06-30
Published:2017-09-07
Contact:
Xue-li ZHENG
E-mail:mzhengxueli2001@126.com
Supported by:CLC Number:
Yu-yan GUO, Lei LUO, Zhang-yao SONG, Xue-li ZHENG. Mitochondrial DNA sequence analysis of cytochrome oxidase subunit Ⅰ gene from 28 mosquito species in China[J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2017, 35(3): 280-287.
Table 1
The collection sites, species and number of mosquitoes
| 采集地 Collection sites | 蚊种 (数量/只) Species (Number) | ||
|---|---|---|---|
| 广东省Guangdong Province | 广州Guangzhou | 23°07′N 113°15′E | 白纹伊蚊 Ae. albopictus (10),致倦库蚊 Cx. quinquefasciatus (2),褐尾库蚊 Cx. fuscanus (1) |
| 深圳Shenzhen | 22°32′N 113°55′E | 白纹伊蚊 Ae. albopictus (10),致倦库蚊 Cx. quinquefasciatus (2) | |
| 东莞Dongguan | 22°28′N 111°24′E | 白纹伊蚊 Ae. albopictus (3),致倦库蚊 Cx. quinquefasciatus (1) | |
| 汕头Shantou | 23°21′N 116°40′E | 白纹伊蚊 Ae. albopictus (10),刺扰伊蚊 Ae. vexans (3),三带喙库蚊 Cx. tritaeniorhynchus (1) | |
| 韶关Shaoguan | 24°11′N 113°24′E | 白纹伊蚊 Ae. albopictus (1),三带喙库蚊 Cx. tritaeniorhynchus (1),致倦库蚊Cx. quinquefasciatus (1) | |
| 实验室Lab | 23°07′N 113°15′E | 埃及伊蚊 Ae. aegypti (6) | |
| 广西壮族自治区 Guangxi Zhuang Autonomous Region | 梧州Wuzhou | 23°53′N 110°32′E | 白纹伊蚊 Ae. albopictus (10),中华按蚊 An. sinensis (2),三带喙库蚊 Cx. tritaeniorhynchus (1),致倦库蚊 Cx. quinquefasciatus (1),尖音库蚊 Cx. pipiens (1) |
| 海南省Hainan Province | 海口Haikou | 20°02′N 110°11′E | 刺扰伊蚊 Ae. vexans(1),白纹伊蚊 Ae. albopictus (10),致倦库蚊 Cx.quinquefasciatus (1),骚扰阿蚊 Ar. subalbatus (1) |
| 澄迈Chengmai | 19°44′N 110°00′E | 刺扰伊蚊 Ae. vexans (2),白纹伊蚊 Ae. albopictus (10),嗜人按蚊 An. anthropophagus (1),须喙按蚊 An. barbirostis (1),中华按蚊 An. sinensis (1),棋斑按蚊 An. tessellatus (1),迷走按蚊 An. vagus(1),褐尾库蚊 Cx. fuscanus (1),三带喙库蚊 Cx. tritaeniorhynchus(1),致倦库蚊 Cx.quinquefasciatus (1),骚扰阿蚊 Ar. subalbatus (1),华丽巨蚊 Tx. splendens (1),杂鳞库蚊 Cx. vishnui (3),常型曼蚊 Ma. uniformis (1) | |
| 儋州Danzhou | 19°31′N 109°34′E | 白纹伊蚊 Ae. albopictus (10),三带喙库蚊 Cx. tritaeniorhynchus(1),致倦库蚊 Cx. quinquefasciatus (1),白雪库蚊 Cx. gelidus (1) | |
| 琼海Qionghai | 19°10′N 110°22′E | 白纹伊蚊 Ae. albopictus(8),白跗按蚊 An. albitarsis(1),中华按蚊 An. sinensis (1),棋斑按蚊 An. tessellatus (2),迷走按蚊 An. vagus (1),三带喙库蚊 Cx. tritaeniorhynchus(1),棕头库蚊 Cx. fuscocehala(1),白胸库蚊 Cx. pallidothorax (2),致倦库蚊 Cx. quinquefasciatus (1) | |
| 白沙Baisha | 19°13′N 109°26′E | 刺扰伊蚊 Ae. vexans (1),窄翅伊蚊 Ae. lineatopennis (1),白纹伊蚊 Ae. albopictus (10),嗜人按蚊 An. anthropophagus (2),须喙按蚊 An. barbirostis (2),中华按蚊 An. sinensis (1),棋斑库蚊 An. tessellatus(1),三带喙库蚊 Cx. tritaeniorhynchus (2),带纹库蚊 Cx. cinctellus (1),白雪库蚊 Cx. gelidus (1),幼小库蚊 Cx. infantulus (1),薛氏库蚊 Cx. shebbearei (2),骚扰阿蚊 Ar.subalbatus (1) | |
| 琼中Qiongzhong | 19°02′N 109°50′E | 窄翅伊蚊 Ae. lineatopennis (2),白纹伊蚊 Ae. albopictus (10),白胸库蚊 Cx. pallidothorax (3),骚扰阿蚊 Ar. subalbatus (1) | |
| 昌江Changjiang | 19°17′N 109°03′E | 刺扰伊蚊 Ae. vexans (1),白纹伊蚊 Ae. albopictus (10),棋斑按蚊 An. tessellatus (1),三带喙库蚊 Cx. tritaeniorhynchus (5),致倦库蚊 Cx.quinquefasciatus(1),杂鳞库蚊 Cx. vishnui (1),骚扰阿蚊 Ar. subalbatus (2) | |
| 陵水Lingshui | 18°30′N 110°01′E | 窄翅伊蚊 Ae. lineatopennis (1),白纹伊蚊 Ae. albopictus (10),白跗按蚊 An. albitarsis (1),嗜人按蚊 An. anthropophagus (1),棋斑按蚊 An. tessellatus (1),迷走按蚊 An. vagus (5),三带喙库蚊 Cx. tritaeniorhynchus (3),致倦库蚊 Cx. quinquefasciatus (1),海滨库蚊 Cx. sitiens (2),骚扰阿蚊 Ar. subalbatus (1),常型曼蚊 Ma. uniformis (1) | |
| 保亭Baoting | 18°38′N 109°41′E | 白纹伊蚊 Ae.albopictus (10),三带喙库蚊 Cx. tritaeniorhynchus (1),白胸库蚊Cx. pallidothorax (2),尖音库蚊 Cx. pipiens (1),骚扰阿蚊 Ar. subalbatus (3) | |
| 三亚Sanya | 19°16′N 111°47′E | 中华按蚊 An. sinensis (1),三带喙库蚊 Cx. tritaeniorhynchus (1),致倦库蚊 Cx. quinquefasciatus (1) | |
| 乐东Ledong | 18°45′N 109°14′E | 白纹伊蚊 Ae. albopictus (1),三带喙库蚊 Cx. tritaeniorhynchus (2) | |
| 福建省 Fujian Province | 古田Gutian | 25°13′N 118°15′E | 白纹伊蚊 Ae. albopictus (4),致倦库蚊 Cx. quinquefasciatus (1) |
| 浙江省Zhejiang Province | 金华Jinhua | 28°56′N 119°13′E | 白纹伊蚊 Ae. albopictus (3),中华按蚊 An. sinensis (1),二带喙库蚊 Cx. bitaeniorhynchus (1),致倦库蚊 Cx. quinquefasciatus (1),骚扰阿蚊 Ar. subalbatus (1) |
| 山西省Shanxi Province | 吕梁Lvliang | 34°29′N 113°15′E | 淡色库蚊 Cx. pipiens pallens (1) |
| 河南省Henan Province | 郑州Zhengzhou | 34°38′N 113°43′E | 淡色库蚊 Cx. pipiens pallens (3) |
| 开封Kaifeng | 34°47′N 114°18′E | 窄翅伊蚊 Ae. lineatopennis (1),白纹伊蚊 Ae. albopictus (11),中华按蚊 An. sinensis (2),三带喙库蚊 Cx. tritaeniorhynchus (2) | |
| 天津 Tianjin | 天津Tianjin | 38°56′N 113°12′E | 淡色库蚊 Cx. pipiens pallens (1) |
| 内蒙古自治区 Neimenggu Autonomous Region | 呼伦贝尔 Hulunbeier | 38°39′N 104°04′E | 背点伊蚊 Ae. dorsalis (1) |
| 吉林省Jilin Province | 吉林Jilin | 42°50′N 128°22′E | 淡色库蚊 Cx. pipiens pallens (1) |
| 黑龙江Heilongjiang Province | 齐齐哈尔Qiqihaer | 47°21′N 124°11′E | 淡色库蚊 Cx. pipiens pallens (1) |
| 云南省Yunnan Province | 昆明Kunming | 25°17′N 102°58′E | 三带喙库蚊 Cx. tritaeniorhynchus (1),致倦库蚊 Cx. quinquefasciatus (1) |
| 景洪Jinghong | 22°00′N 100°46′E | 白纹伊蚊 Ae. albopictus (10) | |
| 勐海Menghai | 21°49′N 103°03′E | 三带喙库蚊 Cx. tritaeniorhynchus (1) | |

Fig. 1
Gel electrophoresis results of amplified products of COⅠ gene from 28 mosquito species M: DNA marker; 1-28: Ae. albopictus, Ae. aegypti, Ae. vexans, Ae. lineatopennis, Ae. dorsalis, An. sinensis, An. anthropophagus, An. tessellatus, An. vagus, An. barbirostris, An. albitarsis, Cx. fuscanus, Cx. bitaeniorhynchus, Cx. tritaeniorhynchus, Cx. pipiens quinquefasciatus, Cx. pipiens pallens, Cx. pipiens, Cx. sitiens, Cx. cinctellus, Cx. fuscocephala, Cx. gelidus, Cx. pallidothorax, Cx. infantulus, Cx. shebbearei, Cx. vishnui, Ar. subalbatus, Ma. uniformis and Tx. splendens.
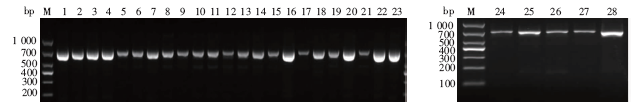
Table 2
The intraspecies and interspecies genetic distances of 6 species of Anopheles
| 蚊种Species | 白跗按蚊 An. albitarsis | 嗜人按蚊 An. anthropophagus | 须喙按蚊 An. barbirostis | 中华按蚊 An. sinensis | 迷走按蚊 An. vagus | 棋斑按蚊 An. tessellatus |
|---|---|---|---|---|---|---|
| 白跗按蚊 An. albitarsis | 0.015 6 | 0.010 6 | 0.004 1 | 0.011 7 | 0.012 6 | 0.005 5 |
| 嗜人按蚊An. anthropophagus | 0.145 0 | |||||
| 须喙按蚊An. barbirostis | 0.138 8 | 0.105 6 | ||||
| 中华按蚊An. sinensis | 0.143 2 | 0.010 7 | 0.103 7 | |||
| 迷走按蚊An. vagus | 0.015 8 | 0.145 1 | 0.139 9 | 0.142 4 | ||
| 棋斑按蚊An. tessellatus | 0.131 0 | 0.107 2 | 0.090 4 | 0.106 4 | 0.130 5 |
|
| [1] | JIANG Ning, BAI Jie, LI Ping, SHAN Wenqi, ZHOU Qiuming, DONG Haowei, YUAN Hao, TAO Feng, LI Xiangyu, MA Yajun, PENG Heng. Establishment of a rapid detection method of mosquito-borne dengue virus based on loop-mediated isothermal amplification microfluidic chip technology [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(2): 234-241. |
| [2] | ZHANG Hong, CHENG Gong. Research progress on the effect of human volatiles on mosquito behavior [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(1): 10-14. |
| [3] | JIANG Li, ZHANG Yao-guang, LIU Hong-xia, WANG Zhen-yu, ZHU Min, WU Huan-yu. Establishment of multiplex PCR for malaria-transmitting vector surveillance [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(2): 159-167. |
| [4] | GUO Hong-xia, ZHAO Teng, WU Jia-hong, LI Chun-xiao. Research progress on the effects of human skin microbiota on mosquito olfactory behavior [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(1): 94-98. |
| [5] | WAN Lun, ZHANG Hua-xun, LI Kai-jie, ZHANG Cong, CAO Mu-min, WU Dong-ni, ZHANG Juan, LIN Wen, LIU Si, ZHU Hong, XIA Jing. Surveillance of malaria-transmitting vectors in Hubei Province from 2018 to 2020 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(5): 592-597. |
| [6] | CHEN Sui-lin, LIU Tai-ping, XU Wen-yue. Development of malaria vaccines and the challenges [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(3): 283-295. |
| [7] | LIU Tong, WU Yang, LIU Pei-wen, YANG Wen-qiang, JIN Bin-bin, GU Jin-bao, CHEN Xiao-guang. Olfactory perception and sex determination of vector mosquitoes [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(1): 1-7. |
| [8] | GU Zhen-yu, ZHAO Teng, LI Chun-xiao. Research progress on odor binding proteins and odor receptors of mosquitoes [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(6): 753-757. |
| [9] | WANG Yang, LI Ting-ting, GONG Mao-qing. Advances in research on olfactory receptors of mosquitoes [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(5): 647-652. |
| [10] | YE Sheng-yu, CHENG Yi-yi, LI Man, ZHOU Hong-ning. Advances in methods for detecting drug-resistance molecular markers of Plasmodium falciparum [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(4): 490-495. |
| [11] | LIU Jian-xiu, WANG Jun-yun. Applications of molecular techniques in classification and identification of Leishmania [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(4): 508-513. |
| [12] | Zhen CAI, Xi YU, Gong CHENG. Progress towards mosquito microbiome on regulating the transmission of mosquito-borne diseases [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2019, 37(5): 603-608. |
| [13] | Xiao-ming WANG, Kun WU, Xiao-guang CHEN, Gui-yun YAN. Research advances on diversity and function of mosquito-bacteria symbiosis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2017, 35(3): 305-312. |
| [14] | Yu-yan GUO, Lei LUO, Xue-li ZHENG. Research progress on application of DNA barcoding technique in Culicidae taxonomy [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2017, 35(1): 93-98. |
| [15] | XU Chao, WEI Qing-kuan,LI Jin,XIAO Ting,YIN Kun,WANG Yong-bin,KONG Xiang-li,XU Yan, CUI Yong,SUN Hui,ZHU Song,YAN Ge,HUANG Bing-cheng*. Detection of Genetic Mutations Associated with Drug Resistance in Imported Plasmodium falciparum [J]. , 2016, 34(6): 2-482-488. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||