
CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES ›› 2024, Vol. 42 ›› Issue (1): 69-77.doi: 10.12140/j.issn.1000-7423.2024.01.010
• ORIGINAL ARTICLES • Previous Articles Next Articles
QIAO Tingting( ), TAO Xianglin, YE Changjiang, LI Zheng, ZHOU Xiaoyan, SUN Entao*(
), TAO Xianglin, YE Changjiang, LI Zheng, ZHOU Xiaoyan, SUN Entao*( )
)
Received:2023-09-22
Revised:2024-01-10
Online:2024-02-28
Published:2024-03-12
Contact:
*E-mail: Supported by:CLC Number:
QIAO Tingting, TAO Xianglin, YE Changjiang, LI Zheng, ZHOU Xiaoyan, SUN Entao. Analysis on genetic variation and differentiation of Tyrophagus putrescentiae in different geographic populations[J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2024, 42(1): 69-77.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.jsczz.cn/EN/10.12140/j.issn.1000-7423.2024.01.010
Table 1
Collection information of T. putrescentiae samples
| 采集地 Location | 代码 Code | 采集日期 Date of sampling | 经纬度 Longitude and latitude | 标本数量 No. samples |
|---|---|---|---|---|
| 安徽芜湖Wuhu City, Anhui Province | WH | 2018年6月 June, 2018 | 31.20°N,118.21°E | 15 |
| 安徽阜阳Fuyang City, Anhui Province | FY | 2018年7月 July, 2018 | 32.90°N,115.82°E | 13 |
| 河北石家庄Shijiazhuang City, Hebei Province | SJZ | 2018年7月 July, 2018 | 38.05°N,114.52°E | 12 |
| 河北张家口Zhangjiakou City, Hebei Province | ZJK | 2018年6月 June, 2018 | 40.82°N,114.88°E | 8 |
Table 2
Analysis of molecular variance of T. putrescentiae population based on Cytb and ITS sequences
| 基因 Gene | 种群 Population | 基因序列号 GenBank Accession No. | 单倍型/样本数 Haplotype/ Sample size | 单倍型及其个数 Haplotype and its number | 单倍型多样性 Haplotype diversity (Hd) | 核苷酸多样性 Nucleotide diversity (Pi) |
|---|---|---|---|---|---|---|
| Cytb | WH | MH715737~MH715746 | 6月10日 | H1(1), H2(1), H3(3), H4(3), H5(1), H6(1) | 0.867 | 0.008 42 |
| FY | MH715747~MH715755 | 4月9日 | H4(6), H7(1), H8(1), H9(1) | 0.583 | 0.001 79 | |
| SJZ | MH715756~MH715763 | 3月8日 | H10(6), H11(1), H12(1) | 0.464 | 0.001 34 | |
| ZJK | MH715764~MH715773 | 4月10日 | H13(6), H14(1), H15(2), H16(1) | 0.644 | 0.108 90 | |
| ITS | WH | MH793968~MH793974 | 7月7日 | G1(1), G2(1), G3(1), G4(1), G5(1), G6(1), G7(1) | 1 | 0.390 66 |
| FY | MH793975~MH793984 | 10月10日 | G8(1), G9(1), G10(1), G11(1), G12(1), G13(1), G14(1), G15(1), G16(1), G17(1) | 1 | 0.434 40 | |
| SJZ | MH793985~MH793994 | 10月10日 | G18(1), G19(1), G20(1), G21(1), G22(1), G23(1), G24(1), G25(1), G26(1), G27(1) | 1 | 0.454 54 | |
| ZJK | MH793995~MH793999 | 5月5日 | G28(1), G29(1), G30(1), G31(1), G32(1) | 1 | 0.488 14 |
Table 3
Genetic diversity of T. putrescentiae based on Cytb and ITS sequences
| 基因 Gene | 遗传差异来源 Source of genetic variation | 自由度 Degree of freedom | 平方和 Sum of square | 变异组分 Variance component | 变异百分比/% Percentage of variation/% | Fst值 Fst value |
|---|---|---|---|---|---|---|
| Cytb | 种群间 Inter-population | 3 | 68.967 | 1.832 33 | 23.14 | 0.231 42a |
| 种群内 Intra-population | 33 | 200.817 | 6.085 35 | 78.86 | ||
| 合计 Total | 36 | 269.784 | 7.917 69 | 100 | ||
| ITS | 种群间 Inter-population | 3 | 3 083.412 | 96.381 49 | 25.96 | 0.259 64a |
| 种群内 Intra-population | 28 | 7 695.057 | 274.823 47 | 74.04 | ||
| 合计 Total | 31 | 10 778.469 | 371.204 96 | 100 |
Table 4
Genetic differentiation index and gene flow of T. putrescentiae populations based on Cytb and ITS sequences
| 基因 Gene | 地理种群 Geographical population | 地理种群 Geographical population | |||
|---|---|---|---|---|---|
| WH | FY | SJZ | ZJK | ||
| Cytb | WH | - | 0.747 84 | 0.661 14 | 2.550 65 |
| FY | 0.400 69a | - | 0.058 90 | 3.437 05 | |
| SJZ | 0.430 61a | 0.894 61a | - | 1.622 89 | |
| ZJK | 0.163 90a | 0.127 00 | 0.235 53a | - | |
| ITS | WH | - | 1.697 44 | 1.428 56 | 0.679 08 |
| FY | 0.227 54a | - | inf | 0.771 05 | |
| SJZ | 0.259 26a | -0.017 52 | - | 0.831 48 | |
| ZJK | 0.424 06a | 0.393 37a | 0.375 52a | - | |
Table 5
Results of T. putrescentiae geographical populations neutral test based on Cytb and ITS sequences
| 基因 Gene | 种群 Population | Tajima’s D值 Tajima’s D value | P值 P value | Fu’s FS值 Fu’s FS value | P值 P value |
|---|---|---|---|---|---|
| Cytb | WH | -0.066 12 | 0.493 | -0.711 58 | 0.262 |
| FY | -1.512 97 | 0.044 | -1.891 65 | 0.016 | |
| SJZ | -1.310 09 | 0.094 | -0.998 99 | 0.056 | |
| ZJK | -0.059 71 | 0.526 | 12.947 55 | 1.000 | |
| 合计 Total | -0.737 22 | 0.289 | 2.336 33 | 0.334 | |
| ITS | WH | 0.785 61 | 0.768 | 3.138 3 | 0.625 |
| FY | 2.269 87 | 0.978 | 2.463 84 | 0.675 | |
| SJZ | 2.659 63 | 0.997 | 2.510 93 | 0.757 | |
| ZJK | 2.406 07 | 0.998 | 4.065 11 | 0.659 | |
| 合计 Total | 2.030 29 | 0.935 25 | 3.044 54 | 0.679 |
Fig. 2
Haplotype network diagram of T. putrescentiae based on Cytb gene sequence Note: There are four populations based on geographic distribution, with each colour representing a population. The population classification is the same as Table 1. Each haplotype is represented by a circle, where the size of the circle is proportional to the frequency of the haplotype.Branch lengths of more than one mutation step between haplotypes are labelled with a number, others are one mutation step.
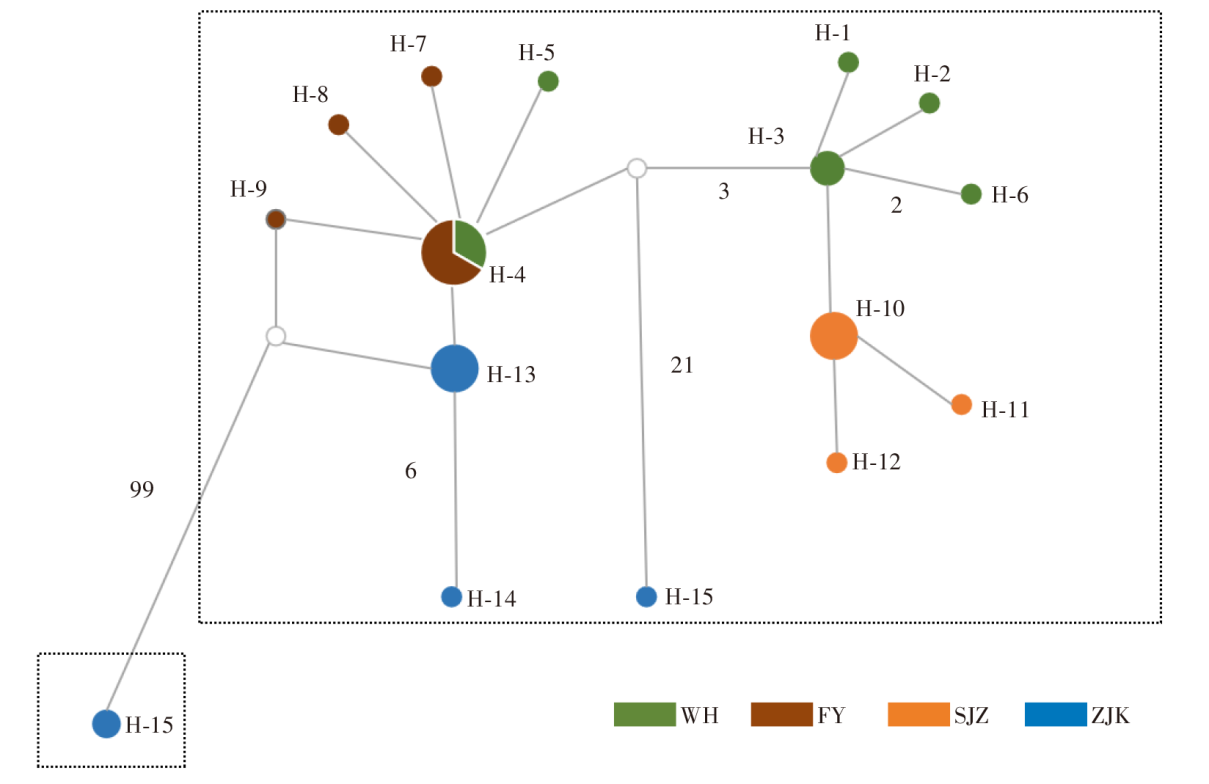
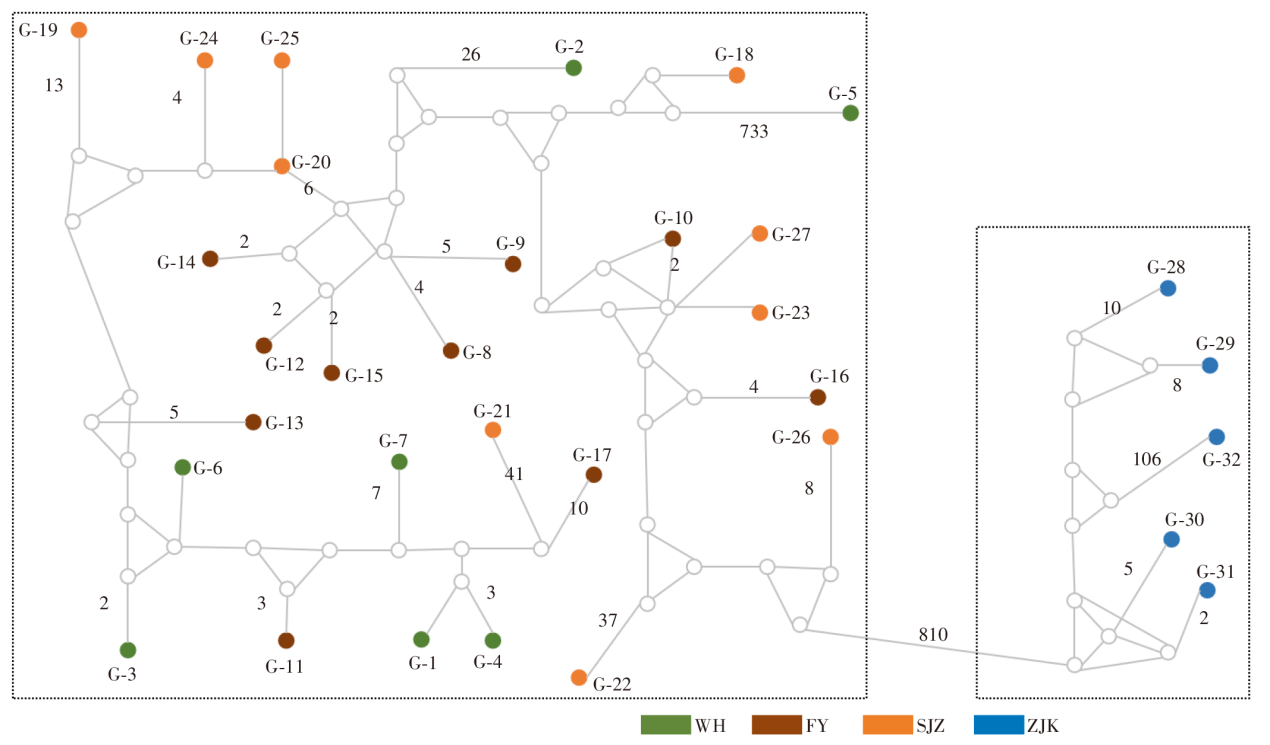
Fig. 4
Haplotype network diagram of T. putrescentiae based on ITS sequence Note: There are 4 population clusters based on geographic distribution, with each colour representing a population. The population classification is the same as Table 1. Each haplotype is represented by a circle, where the size of the circle is proportional to the frequency of the haplotype.Branch lengths of more than one mutation step between haplotypes are labelled with a number. Others are one mutation step.
| [1] | Guo W, Jiang YX. Morphological observation on adult Tyrophagus putrescentiae[J]. J Biol, 2014, 31(3): 95-98. (in Chinese) |
| (郭伟, 姜玉新. 腐食酪螨成螨形态观察[J]. 生物学杂志, 2014, 31(3): 95-98.) | |
| [2] | Yu X, Fan QH. Occurrence and control of Tyrophagus putrescentiae[J]. Fujian Agric Sci Technol, 2002(6): 49-50. (in Chinese) |
| (于晓, 范青海. 腐食酪螨的发生与防治[J]. 福建农业科技, 2002(6): 49-50.) | |
| [3] | Hong Y, Chai Q, Tao N, et al. A case of dermatitis caused by Tyrophagus putrescentiae[J]. Chin J Schisto Control, 2017, 29(3): 395-396. (in Chinese) |
| (洪勇, 柴强, 陶宁, 等. 腐食酪螨致皮炎1例[J]. 中国血吸虫病防治杂志, 2017, 29(3): 395-396.) | |
| [4] | Tao N, Zhan XD, Sun ET, et al. Investigation of Acaroid mites breeding in stored dry fruits[J]. Chin J Schisto Control, 2015, 27(6): 634-637. (in Chinese) |
| (陶宁, 湛孝东, 孙恩涛, 等. 储藏干果粉螨污染调查[J]. 中国血吸虫病防治杂志, 2015, 27(6): 634-637.) | |
| [5] |
Li CP, Cui YB, Wang J, et al. Acaroid mite, intestinal and urinary acariasis[J]. World J Gastroenterol, 2003, 9(4): 874-877.
doi: 10.3748/wjg.v9.i4.874 |
| [6] | Zhan XD. Temporal and spatial patterns of population genetic differentiation of Tyrophagus putrescentiae[D]. Wuhu: Anhui Normal University, 2019. (in Chinese) |
| (湛孝东. 腐食酪螨种群遗传分化的时空格局[D]. 芜湖: 安徽师范大学, 2019.) | |
| [7] | Zhang YC, Lu SH, Gong DF, et al. Analysis of genetic diversity and genetic structure in geographic populations of Corythucha ciliata (Hemiptera ∶ Tingidae) from China based on mitochondrial DNA COⅠgene sequences[J]. Sci Silvae Sin, 2021, 57(8): 102-111. (in Chinese) |
| 张元臣, 卢绍辉, 龚东风, 等. 基于COⅠ基因的我国悬铃木方翅网蝽(半翅目: 网蝽科)种群遗传多样性和遗传结构分析[J]. 林业科学, 2021, 57(8): 102-111.) | |
| [8] |
Wei DD, Yuan ML, Wang BJ, et al. Population genetics of two asexually and sexually reproducing psocids species inferred by the analysis of mitochondrial and nuclear DNA sequences[J]. PLoS One, 2012, 7(3): e33883.
doi: 10.1371/journal.pone.0033883 |
| [9] | Ye CJ, Wang Y, Li XM, et al. Population genetic structure of Dermatophagoides farinae (Acari∶Pyroglyphidae) from different geographic populations based on mitochondrial cytochrome b gene[J]. Saa, 2023, 28(7): 1224-1234. |
| [10] |
Queiroz MCV, Douin M, Sato ME, et al. Molecular variation of the cytochrome b DNA and protein sequences in Phytoseiulus macropilis and P. persimilis (Acari∶Phytoseiidae) reflect population differentiation[J]. Exp Appl Acarol, 2021, 84(4): 687-701.
doi: 10.1007/s10493-021-00648-w |
| [11] |
Guzman-Valencia S, Santillán-Galicia MT, Guzmán-Franco AW, et al. Contrasting effects of geographical separation on the genetic population structure of sympatric species of mites in avocado orchards[J]. Bull Entomol Res, 2014, 104(5): 610-621.
doi: 10.1017/S0007485314000388 |
| [12] | Tao XL, Ma F, Li Z, et al. Genetic variations in four geographical isolates of Gohieria fusca based on cytochrome b and internal transcribed spacer genes[J]. Chin J Schisto Control, 2023, 35(1): 22-28. (in Chinese) |
| (陶香林, 马飞, 李政, 等. 基于细胞色素b和内转录间隔区基因序列的4个棕脊足螨地理种群遗传变异分析[J]. 中国血吸虫病防治杂志, 2023, 35(1): 22-28.) | |
| [13] | Zhang T. Studies on the Population Ecology of Tyrophagus putrescentiae (Schrank) (Acari∶Acaridae)[D]. Nanchang: Nanchang University, 2007. (in Chinese) |
| (张涛. 腐食酪螨种群生态学研究[D]. 南昌: 南昌大学, 2007 ) | |
| [14] | Zheng LX, Yin CC, Wang YX, et al. Identification of Acaroid mite species based on DNA barcoding[J]. Chin J Vector Biol Control, 2019, 30(2): 180-184. (in Chinese) |
|
(郑凌霄, 尹灿灿, 王逸枭, 等. 基于DNA条形码技术的粉螨种类鉴定研究[J]. 中国媒介生物学及控制杂志, 2019, 30(2): 180-184.)
doi: 10.11853/j.issn.1003.8280.2019.02.015 |
|
| [15] | Zheng LX. Molecular identification and phylogeny of acaroid mites[D]. Wuhu: Wannan Medical College, 2019. (in Chinese) |
| (郑凌霄. 常见储藏物粉螨的分子鉴定与系统发育研究[D]. 芜湖: 皖南医学院, 2019.) | |
| [16] |
Dermauw W, van Leeuwen T, Vanholme B, et al. The complete mitochondrial genome of the house dust mite Dermatophagoides pteronyssinus (Trouessart): a novel gene arrangement among arthropods[J]. BMC Genomics, 2009, 10: 107.
doi: 10.1186/1471-2164-10-107 pmid: 19284646 |
| [17] |
Sun ET, Li CP, Nie LW, et al. The complete mitochondrial genome of the brown leg mite, Aleuroglyphus ovatus (Acari∶Sarcoptiformes): evaluation of largest non-coding region and unique tRNAs[J]. Exp Appl Acarol, 2014, 64(2): 141-157.
doi: 10.1007/s10493-014-9816-9 |
| [18] |
Thompson JD, Gibson TJ, Plewniak F, et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools[J]. Nucl Acid Res, 1997, 25(24): 4876-4882.
doi: 10.1093/nar/25.24.4876 |
| [19] |
Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data[J]. Bioinformatics, 2009, 25(11): 1451-1452.
doi: 10.1093/bioinformatics/btp187 pmid: 19346325 |
| [20] |
Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Mol Biol Evol, 2018, 35(6): 1547-1549.
doi: 10.1093/molbev/msy096 pmid: 29722887 |
| [21] |
Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis[J]. Evol Bioinform Online, 2007, 1: 47-50.
pmid: 19325852 |
| [22] |
Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies[J]. Mol Biol Evol, 1999, 16(1): 37-48.
doi: 10.1093/oxfordjournals.molbev.a026036 pmid: 10331250 |
| [23] | Zhan XD, Guo W, Chai Q, et al. Study on natural population dynamics and spatial distribution pattern of Ty-rophagus putrescentiae in stored flour[J]. Chin J Schisto Control, 2017, 29(5): 587-591. (in Chinese) |
| (湛孝东, 郭伟, 柴强, 等. 仓储面粉中腐食酪螨自然种群消长动态及空间分布型研究[J]. 中国血吸虫病防治杂志, 2017, 29(5): 587-591.) | |
| [24] |
Zhang N, Smith CL, Yin Z, et al. Effects of temperature on the adults and progeny of the predaceous mite Lasioseius japonicus (Acari∶Blattisociidae) fed on the cereal mite Tyrophagus putrescentiae (Acari ∶ Acaridae)[J]. Exp Appl Acarol, 2022, 86(4): 499-515.
doi: 10.1007/s10493-022-00708-9 pmid: 35389177 |
| [25] | Xu R. Genetic diversity in populations of Tyrophagus putrescentiae (Schrank) revealed by inter simple sequence repeats (ISSR)[D]. Nanchang: Nanchang University, 2007. (in Chinese) |
| (许睿. 运用ISSR分子标记对腐食酪螨不同种群遗传多样性的研究[D]. 南昌: 南昌大学, 2007.) | |
| [26] |
Tsagkarakou A, Navajas M, Papaioannou-Souliotis P, et al. Gene flow among Tetranychus urticae (Acari ∶ Tetranychidae) populations in Greece[J]. Mol Ecol, 1998, 7(1): 71-79.
doi: 10.1046/j.1365-294x.1998.00305.x |
| [27] |
Ma QZ, He K, Wang XD, et al. Better resolution for cytochrome b than cytochrome c oxidase subunit I to identify Schizothorax species (Teleostei∶Cyprinidae) from the Tibetan Plateau and its adjacent area[J]. DNA Cell Biol, 2020, 39(4): 579-598.
doi: 10.1089/dna.2019.5031 |
| [28] | Liu YY, Yao LS, Ci Y, et al. Genetic differentiation of geographic populations of Rattus tanezumi based on the mitochondrial Cytb gene[J]. PLoS One, 2021, 16(3): e0248102. |
| [29] |
Li XM, Tao XL, Wang Y, et al. Population genetics of Lepidoglyphus destructor inferred by the analysis of the mitochondrial cytochrome b gene and ribosomal internal transcribed spacer gene sequence[J]. Int J Acarol, 2021, 47(8): 670-676.
doi: 10.1080/01647954.2021.1985604 |
| [30] |
Palyvos NE, Emmanouel NG, Saitanis CJ. Mites associated with stored products in Greece[J]. Exp Appl Acarol, 2008, 44(3): 213-226.
doi: 10.1007/s10493-008-9145-y pmid: 18379887 |
| [31] | Yang XQ. Study on the systematic geographical structure of carnivorous mites in malacca in southeast China based on mitochondrial COI Sequence[D]. Nanchang: Nanchang University, 2013. (in Chinese) |
| (杨小强. 基于线粒体COI序列的中国东南地区马六甲肉食螨的系统地理结构研究[D]. 南昌: 南昌大学, 2013.) |
| [1] | MA Zhiya, XIE Shichen, HE Yuanhui, GAO Wenwei, LIU Qing, ZHU Xingquan, ZHENG Wenbin. Infection status and genetic variation analysis of Haemonchus contortus in sheep in Shanxi Province [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(6): 733-738. |
| [2] | LI Yuqiong, YU Youli, GAO Junrong, LIU Yunyun, LI Hongbing, NIU Xiaohao. Enterocytozoon bieneusi infection in dairy cows and its genotype identification in Yinchuan area of Ningxia Province [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(4): 476-479. |
| [3] | HU Kun-min, CHEN Shao-hong, AI Lin, ZHENG Bin. Sequence analysis of ribosomal ITS2 gene and mitochondrial CO1 gene of Paragonimus metacercariae from freshwater crabs in Henan, Anhui, Fujian and Zhejiang provinces, China [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(1): 87-94. |
| [4] | Fang-zhou CHENG, Chun-hua GAO, Yue-tao YANG, Jun-yun WANG. Polymorphism analysis of internal transcribed spacer 1 of Leishmania isolates from different endemic areas in China [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2019, 37(2): 196-201. |
| [5] | Meng XU, Wen HUANG, Shen OU, Jian-hai YIN, Sheng-kui CAO, Li-yu MENG, Jian-ping CAO, Yu-juan SHEN. Infection status and molecular identification of Clonorchis sinensis in human population of Tengxian County, Guangxi [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2019, 37(1): 28-32. |
| [6] | Yue HU, Yan CHEN, Jin-fu LI, Xing-hui MENG, Qiao-xia LIU, Jian LIU. Feasibility assessment of using parasitic species-specific genes to detect plerocercoid infection in worm-free pathological tissue of mice [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2018, 36(4): 357-361. |
| [7] | Qi CAI, Nai-fang YE, Lin AI, Jia-xu CHEN, Jian-biao WANG. Clinical features and diagnosis of an imported loiasis patient in Shanghai [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2018, 36(4): 370-374. |
| [8] | Qiu-an HU, Shan LV, Yun-hai GUO, He-xiang LIU, Yi ZHANG. Genetic diversity of Angiostrongylus cantonensis in Nan’ao Island of Guangdong Province [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2017, 35(2): 130-135. |
| [9] | XU Qian-ming*, ZHAO Chang-cheng, FANG Fen, XU Xiao-pei,LIU Wen-ge, Zhang Qin, WANG Gui-jun. Morphological Characteristics and Molecular Identification of Trichuris sp. from Giraffe [J]. , 2016, 34(2): 4-109-112. |
| [10] | CHANGZhen-shan;WUBuo;BlairD;ZHANGYong-nian;HULing;CHENShao-hong;CHENMing-gang;FENGZhen;GeorgeM.Davis. GENE SEQUENCING FOR IDENTIFICATION OF PARAGONIMUS EGGS FROM A HUMAN CASE [J]. , 2000, 18(4): 6-215. |
| [11] | XuXiaochun;XuJiannong;QuFengyi. DIFFERENTIATION OF CRYPTIC SPECIES A AND D OF ANOPHELES DIRUS COMPLEX BY POLYMERASE CHAIN REACTION [J]. , 1998, 16(3): 172-175. |
| [12] | XuJiannong;QuFengyi. INVESTIGATION OF THE SPECIESSTATUSOF ANOPHELES DIRUS( DIPTERA:CULICIDAE) FROM HAINAN PROVINCE USING rDNA [J]. , 1997, 15(5): 269-272. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||