
CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES ›› 2020, Vol. 38 ›› Issue (2): 181-187.doi: 10.12140/j.issn.1000-7423.2020.02.008
• ORIGNAL ARTICLES • Previous Articles Next Articles
Jian-xiu LIU, Chun-hua GAO, Yue-tao YANG, Bin ZHENG, Jun-yun WANG*( )
)
Received:2019-12-02
Online:2020-04-30
Published:2020-05-11
Contact:
Jun-yun WANG
E-mail:wangjunyunwjy@163.com
Supported by:CLC Number:
Jian-xiu LIU, Chun-hua GAO, Yue-tao YANG, Bin ZHENG, Jun-yun WANG. Evaluation of the value of K26 sequence applied in identification of Leishmania isolates in China[J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2020, 38(2): 181-187.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.jsczz.cn/EN/10.12140/j.issn.1000-7423.2020.02.008
Fig. 1
Electrophoresis of K26 amplification products from Leishmania isolates in China and from the K26 international reference isolates M: DNA marker; 1-16: Leishmania isolates 801, KS2, KS6, JIASHI-1, JIASHI-2, JIASHI-5, XJ771, KXG-Xu, KXG-927, KXG-918, KXG-Liu, KXG-65, Hebei-Lv, Shanxi-Yang, SC6 and Cy in China; 17: International reference Leishmania isolate DD8; 18-22: Leishmania isolates 5AS, 2903, 3720, L100 and K27, which do not belong to the L. donovani complex
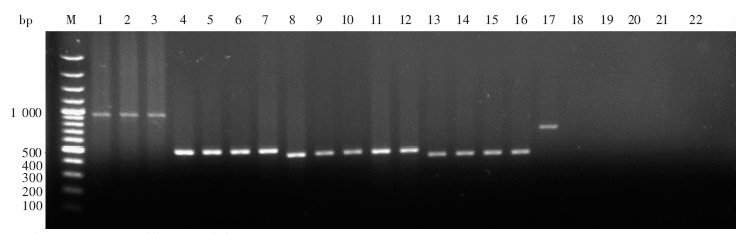
Table 1
The amplified K26 sequence length and GenBank accession numbers of Leishmania isolates in China and international reference isolates
| 流行区 Endemic area | 分离株 Isolate | 世界卫生组织编码 WHO code | 来源 Location | 宿主 Host | 序列长度/bp Sequence length/bp | GenBank登录号 Accession no. |
|---|---|---|---|---|---|---|
| 人源型 Anthroponotic type | 801 | MHOM/CN/1980/801 | 新疆喀什Kashi, Xinjiang | 人Human | 920 | MN702441 |
| KS-2 | MHOM/CN/1996/KS-2 | 新疆喀什Kashi, Xinjiang | 人Human | 920 | MN702442 | |
| KS-6 | MHOM/CN/1996/KS-6 | 新疆喀什Kashi, Xinjiang | 人Human | 920 | MN688111 | |
| 自然疫源型 natural-sourced leishmaniasis | JIASHI-1 | MHOM/CN/2008/JIASHI-1 | 新疆伽师Jiashi, Xinjiang | 人Human | 491 | MN688113 |
| JIASHI-2 | MHOM/CN/2008/JIASHI-2 | 新疆伽师Jiashi, Xinjiang | 人Human | 491 | MN688114 | |
| JIASHI-5 | MHOM/CN/2008/JIASHI-5 | 新疆伽师Jiashi, Xinjiang | 人Human | 491 | MN688115 | |
| XJ771 | IWUI/CN/1977/XJ771 | 新疆巴楚Bachu, Xinjiang | 白蛉Sandfly | 491 | MN688112 | |
| 人犬共患型 zoonosis (specifically canine) leishmaniasis | SC6 | MHOM/CN/1986/SC6 | 四川九寨沟Jiuzhaigou, Sichuan | 人Human | 404 | MN688116 |
| Cy | MCAN/CN/2008/Cy | 甘肃武都Wudu, Gansu | 犬Canine | 404 | MN688117 | |
| Shanxi-Yang | MHOM/CN/2018/Shanxi-Yang | 山西阳泉Yangquan, Shanxi | 人Human | 404 | MN688118 | |
| Hebei-Lv | MHOM/CN/2018/ Hebei-Lv | 河北邯郸Handan, Hebei | 人Human | 404 | MN688119 | |
| 皮肤利什曼病 Cutaneous leishmaniasis | KXG-Xu | MHOM/CN/1994/KXG-Xu | 新疆克拉玛依 Karamay, Xinjiang | 人Human | 449 | MN688120 |
| KXG-927 | IMJW/CN/1992/KXG-927 | 新疆克拉玛依 Karamay, Xinjiang | 白蛉Sandfly | 449 | MN688121 | |
| KXG-918 | IMJW/CN/1991/KXG-918 | 新疆克拉玛依 Karamay, Xinjiang | 白蛉Sandfly | 449 | MN688122 | |
| KXG-65 | IMJW/CN/1987/KXG-65 | 新疆克拉玛依 Karamay, Xinjiang | 白蛉Sandfly | 449 | MN688123 | |
| KXG-Liu | MHOM/CN/1994/KXG-Liu | 新疆克拉玛依 Karamay, Xinjiang | 人Human | 449 | MN688124 | |
| 国际参照株 International reference strains | DD8 | MHOM/IN/1980/DD8 | 印度India | 人Human | 641 | MN702440 |
| 5AS | MHOM/SU/73/5ASKH | 土库曼Turkmenistan | 人Human | - | ||
| 2903 | MHOM/BR/75/M2903 | 巴西Brazil | 人Human | - | ||
| 3720 | MROH/SU/80/CLONE3720 | 乌兹别克Uzbek | 大沙鼠Big gerbils | - | ||
| L100 | MHOM/ET/72/L100 | 埃塞俄比亚Ethiopia | 人Human | - | ||
| K27 | MHOM/SU/74/K27 | 阿塞拜疆Azerbaijan | 人Human | - |
Table 2
Sequence identity of k26 amplification products among Leishmania isolates in China(%)
| 分离株Isolate | 801 | KS-2 | KS-6 | XJ771 | JIASHI-1 | JIASHI-2 | JIASHI-5 | KXG-65 | KXG-918 | KXG-927 | KXG-Liu | KXG-Xu | Cy | Hebei-Lv | Shanxi-Yang | SC6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 801 | 100 | |||||||||||||||
| KS-2 | 100 | 100 | ||||||||||||||
| KS-6 | 100 | 100 | 100 | |||||||||||||
| XJ771 | 49.6 | 49.6 | 49.6 | 100 | ||||||||||||
| JIASHI-1 | 49.6 | 49.6 | 49.6 | 100 | 100 | |||||||||||
| JIASHI-2 | 49.6 | 49.6 | 49.6 | 100 | 100 | 100 | ||||||||||
| JIASHI-5 | 49.6 | 49.6 | 49.6 | 100 | 100 | 100 | 100 | |||||||||
| KXG-65 | 45.2 | 45.2 | 45.2 | 91.4 | 91.4 | 91.4 | 91.4 | 100 | ||||||||
| KXG-918 | 45.2 | 45.2 | 45.2 | 91.4 | 91.4 | 91.4 | 91.4 | 100 | 100 | |||||||
| KXG-927 | 45.2 | 45.2 | 45.2 | 91.4 | 91.4 | 91.4 | 91.4 | 100 | 100 | 100 | ||||||
| KXG-Liu | 45.2 | 45.2 | 45.2 | 91.4 | 91.4 | 91.4 | 91.4 | 100 | 100 | 100 | 100 | |||||
| KXG-Xu | 45.2 | 45.2 | 45.2 | 91.4 | 91.4 | 91.4 | 91.4 | 100 | 100 | 100 | 100 | 100 | ||||
| Cy | 40.2 | 40.2 | 40.2 | 80.2 | 80.2 | 80.2 | 80.2 | 87.8 | 87.8 | 87.8 | 87.8 | 87.8 | 100 | |||
| Hebei-Lv | 40.2 | 40.2 | 40.2 | 80.2 | 80.2 | 80.2 | 80.2 | 87.8 | 87.8 | 87.8 | 87.8 | 87.8 | 100 | 100 | ||
| Shanxi-Yang | 40.2 | 40.2 | 40.2 | 80.2 | 80.2 | 80.2 | 80.2 | 87.8 | 87.8 | 87.8 | 87.8 | 87.8 | 100 | 100 | 100 | |
| SC6 | 40.2 | 40.2 | 40.2 | 80.2 | 80.2 | 80.2 | 80.2 | 87.8 | 87.8 | 87.8 | 87.8 | 87.8 | 100 | 100 | 100 | 100 |
| [1] | Chen JP, Yang BB, Xu JN , et al. Epidemic status of kala-azar in China[ C]//Zoological Society Symposium in Sichuan Province. Chengdu, 2011: 138. (in Chinese) |
| ( 陈建平, 杨斌斌, 徐佳楠 , 等. 我国黑热病流行现状[C]// 四川省动物学会第九次会员代表大会暨第十届学术研讨会论文集. 成都, 2011: 138.) | |
| [2] | Wang JY, Gao CH, Yang YT , et al. An outbreak of the desert sub-type of zoonotic visceral leishmaniasis in Jiashi, Xinjiang Uygur Autonomous Region, People’s Republic of China[J]. Parasitol Int, 2010,59(3):331-337. |
| [3] | Zhou XN, Lv S, Yang GJ , et al. Spatial epidemiology in zoonotic parasitic diseases: insights gained at the 1st International Symposium on Geospatial Health in Lijiang, China, 2007[J]. Parasit Vectors, 2009,2:10. |
| [4] | Guan LR, Qu JQ, Chai JJ . Leishmaniasis in China-present status of prevalence and some suggestions on its control[J]. Endem Dis Bull China, 2000,15(3):49-52. (in Chinese) |
| ( 管立人, 瞿靖琦, 柴君杰 . 中国利什曼病的现状和对开展防治工作的若干建议[J]. 地方病通报, 2000,15(3):49-52.) | |
| [5] | Wang JY, Cui G, Chen HT , et al. Current epidemiological profile and features of visceral leishmaniasis in People’s Republic of China[J]. Parasit Vectors, 2012,5:31. |
| [6] | Guan LR, Yang YQ, Qu JQ , et al. Discovery and study of cutaneous leishmaniasis in Karamay of Xinjiang, West China[J]. Infect Dis Poverty, 2013,2(1):20. |
| [7] | Alce TM, Gokool S ,McGhie D , et al. Expression of hydrophilic surface proteins in infective stages of Leishmania donovani[J]. Mol Biochem Parasitol, 1999,102(1):191-196. |
| [8] | Haralambous C, Antoniou M, Pratlong F , et al. Development of a molecular assay specific for the Leishmania donovani complex that discriminates L. donovani/Leishmania infantum zymodemes: a useful tool for typing MON-1[J]. Diagn Microbiol Infect Dis, 2008,60(1):33-42. |
| [9] | Kimura M . A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences[J]. J Mol Evol, 1980,16(2):111-120. |
| [10] | Saitou N, Nei M . The neighbor-joining method: a new method for reconstructing phylogenetic trees[J]. Mol Biol Evol, 1987,4(4):406-425. |
| [11] | Felsenstein J . Confidence limits on phylogenies: an approach using the bootstrap[J]. Evolution, 1985,39(4):783-791. |
| [12] | Schönian G, Cupolillo E, Mauricio I . Molecular evolution and phylogeny of Leishmania[M] //Drug Resistance in Leishmania Parasites. Vienna: Springer Vienna, 2012: 15-44. |
| [13] | Guan LR, Yang YQ, Zhai JQ , et al. Study of cutaneous leishmaniasis in Karamay, Xinjiang[J]. Chin J Parasitol Parasit Dis, 1997,15(3):181-185. (in Chinese) |
| ( 管立人, 杨元清, 翟靖琦 . 新疆克拉玛依地区皮肤利什曼病的研究[J]. 中国寄生虫学与寄生虫病杂志, 1997,15(3):181-185.) | |
| [14] | Cheng FZ, Gao CH, Yang YT , et al. Polymorphism analysis of internal transcribed spacer 1 of Leishmania isolates from different endemic areas in China[J]. Chin J Parasitol Parasit Dis, 2019,37(2):196-201. (in Chinese) |
| ( 程芳洲, 高春花, 杨玥涛 , 等. 我国不同利什曼病流行区利什曼原虫分离株核糖体内转录间隔区1的多态性分析[J]. 中国寄生虫学与寄生虫病杂志, 2019,37(2):196-201.) | |
| [15] | Bu LY, Hu XS, Jing BQ, Yi TL . Sequence analysis of SSU rDNA variable regions of Leishmania isolates from hillly foci and plain foci of China[J]. Chin J Parasitol Parasit Dis, 2000,18(6):321-324. (in Chinese) |
| ( 卜玲毅, 胡孝素, 敬保迁 , 等. 我国内脏利什曼病山丘疫区与平原疫区利什曼原虫SSUrDNA多变区序列分析[J]. 中国寄生虫学与寄生虫病杂志, 2000,18(6):321-324.) | |
| [16] | Bu LY, Hu XS, Jing BQ , et al. Sequence analysis of SSU rDNA variable region of different Leishmania isolates of China[J]. Pract Parasit Dis, 2001,9(1):1-3. (in Chinese) |
| ( 卜玲毅, 胡孝素, 敬保迁 , 等. 利什曼原虫中国分离株SSU rDNA多变区序列分析[J]. 实用寄生虫病杂志, 2001,9(1):1-3.) | |
| [17] | Yuan DM, Qin HX, Zhang JG , et al. Phylogenetic analysis of HSP70 and cyt b gene sequences for Chinese Leishmania isolates and ultrastructural characteristics of Chinese Leishmania sp[J]. Parasitol Res, 2017,116(2):693-702. |
| [18] | Cao DP, Guo XG, Chen DL , et al. Species delimitation and phylogenetic relationships of Chinese Leishmania isolates reexamined using kinetoplast cytochrome oxidase Ⅱ gene sequences[J]. Parasitol Res, 2011,109(1):163-173. |
| [19] | Guan W, Cao DP, Sun K , et al. Phylogenic analysis of Chinese Leishmania isolates based on small subunit ribosomal RNA (SSU rRNA) and 7 spliced leader RNA (7SL RNA)[J]. Acta parasitologica, 2012,57(2):101-113. |
| [1] | WANG Xiaojun, CAI Yucheng, ZOU Xuan, LI Hui, TONG Bobo. The epidemiological characteristics of visceral leishmaniasis in Longnan City from 2005 to 2021 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(5): 579-585. |
| [2] | ZHANG Li, YI Boyu, YIN Jianhai, XIA Zhigui. Epidemiological characteristics of malaria in China, 2022 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(2): 137-141. |
| [3] | KUI Yan, XUE Chuizhao, WANG Xu, LIU Baixue, WANG Ying, WANG Liying, YANG Shijie, HAN Shuai, WU Weiping, XIAO Ning. Progress of echinococcosis control in China, 2021 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(2): 142-148. |
| [4] | ZHOU Zhengbin, PAN Gaiqin, LI Yuanyuan, LIU Qin, YANG Limin, LI Zhongqiu, MA Zhitao, ZHANG Yi, LI Shizhu. Prevalence of visceral leishmaniasis in China in 2021 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(2): 149-155. |
| [5] | WANG Fenfen, ZHANG Peijun, REN Mengzhi, LI Daohao. Epidemiological characteristics of visceral leishmaniasis in Yangquan City, Shanxi Province from 2006 to 2021 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(2): 228-232. |
| [6] | JIA Zhenzhen, LIU Hongying, JIANG Qi, WANG Lingling, LIU Xiangjun. A case of visceral leishmaniasis misdiagnosed as a hematological disorder [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(2): 257-259. |
| [7] | GU Yan, YU Jin, XU Cehua. Investigation on the first imported visceral leishmaniasis case in Ningxia in the past 44 years [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(2): 260-262. |
| [8] | JIANG Tiange, ZENG Wenbo, LI Zhongqiu, ZHANG Yi. Research advances in the regulatory role of non-coding RNA in leishmaniasis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2023, 41(1): 92-97. |
| [9] | YANG Cheng-yun, HE Zhi-quan, LU De-ling, QIAN Dan, LIU Ying, LI Su-hua, ZHOU Rui-min, DENG Yan, ZHANG Hong-wei, WANG Hao, ZHAO Dong-yang, GUO Wan-shen. Epidemiological investigation on cases of visceral leishmaniasis in Henan Province in 2020 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(4): 481-486. |
| [10] | LI Zhen, GUO Xiao-kui, WANG Yue-xiang, ZHENG Bin, ZHOU Xiao-nong. The strategy on One Health development in China based on SWOT analysis [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(3): 271-277. |
| [11] | CHEN Yuan-cai, HUANG Jian-ying, LI Jun-qiang, ZHANG Long-xian. Molecular epidemiology and subtype distribution of Cryptosporidium parvum from dairy cattle in China [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(3): 278-284. |
| [12] | ZHANG Li, YI Bo-yu, XIA Zhi-gui, YIN Jian-hai. Epidemiological characteristics of malaria in China, 2021 [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(2): 135-139. |
| [13] | LUO Zhuo-wei, ZHOU Zheng-bin, GONG Yan-feng, FENG Jia-xin, LI Yuan-yuan, ZHANG Yi, LI Shi-zhu. Current status and challenges of visceral leishmaniasis in China [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(2): 146-152. |
| [14] | ZHANG Ai-ping, LIANG Man-man, ZHU Ling-ling, SHENG Hao-yu, YANG Jiang-hua. Relapse of visceral leishmaniasis in a cured case after 64 years [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(2): 266-268. |
| [15] | XIA Zhi-gui, FENG Jun, ZHANG Li, FENG Xin-yu, HUANG Fang, YIN Jian-hai, ZHOU Shui-sen, ZHOU Sheng, YANG Heng-lin, WANG Shan-qing, GAO Qi, TANG Lin-hua, YAN Jun. Achieving malaria elimination in China: analysis on implementation and effectiveness of the surveillance-response system [J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2021, 39(6): 733-740. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||