
CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES ›› 2022, Vol. 40 ›› Issue (2): 194-203.doi: 10.12140/j.issn.1000-7423.2022.02.010
• ORIGINAL ARTICLES • Previous Articles Next Articles
SUN Jia-ning( ), CHEN Ting, DONG Wen-ge*(
), CHEN Ting, DONG Wen-ge*( )
)
Received:2021-09-22
Revised:2021-11-18
Online:2022-04-30
Published:2022-04-12
Contact:
DONG Wen-ge
E-mail:954343981@qq.com;dongwenge2740@sina.com
Supported by:CLC Number:
SUN Jia-ning, CHEN Ting, DONG Wen-ge. Sequencing and analysis of the mitochondrial genome of Hoplopleura edentula[J]. CHINESE JOURNAL OF PARASITOLOGY AND PARASITIC DISEASES, 2022, 40(2): 194-203.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.jsczz.cn/EN/10.12140/j.issn.1000-7423.2022.02.010
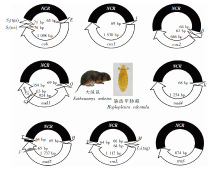
Fig. 1
The fragmented mitochondrial genomes of H. edentula on E. miletus NCR: Non-coding regions; S2(tga): tRNA-Ser (anticodon UGA); S1(tct): tRNA-Ser (anticodon UCU); cob: Cytochrome b; cox1: Cytochrome oxidate subunitⅠ; nad1: NADH dehydrogenase subunit 1; rrnS: Small ribosomal submit RNA; rrnL: Large ribosomal submit RNA; E: tRNA-Glu; I: tRNA-Ile; D: tRNA-Asp; G: tRNA-Gly; Q: tRNA-Gln; K: tRNA-Lys; F: tRNA-Phe; H: tRNA-His; V: tRNA-Val; M: tRNA-Met; T: tRNA-Thr; L1(tag): tRNA-Leu (anticodon UAG).
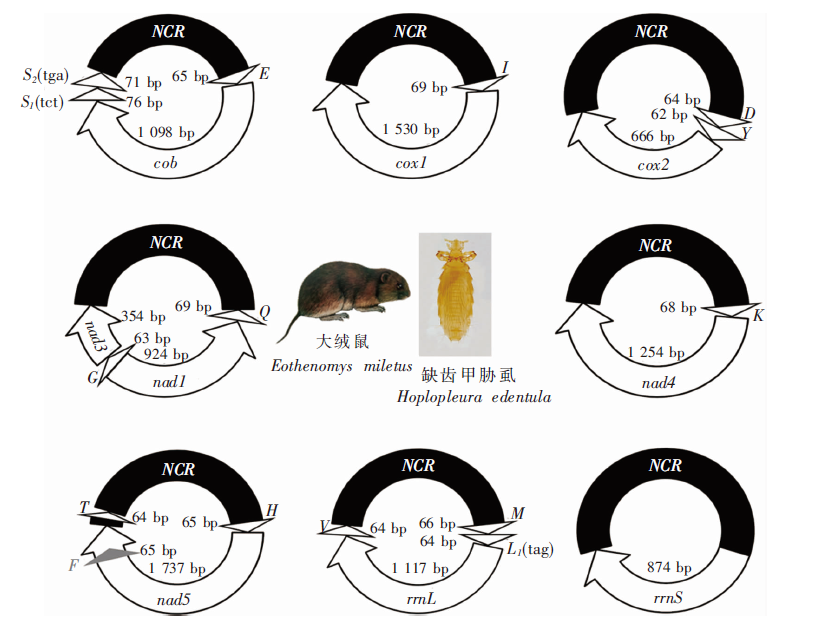
Table 1
The size of minichromosomes coding region of H. edentula on E. miletus
| 微环染色体 Minichromosome | 编码区大小/bp Size of coding region/bp | 序列读数/bp No. sequence read/bp |
|---|---|---|
| E-cob-S1(tct)-S2(tga) | 1 303 | 456 433 |
| I-cox1 | 1 599 | 206 917 |
| D-Y-cox2 | 791 | 822 133 |
| Q-nad1-G-nad3 | 1 446 | 712 883 |
| K-nad4 | 1 322 | 960 072 |
| H-nad5-F-T | 1 889 | 441 440 |
| M-L1(tag)-rrnL-V | 1 311 | 1 681 291 |
| rrnS | 874 | 1 290 852 |
| 合计Total | 10 535 | 6 572 021 |
Table 2
Comparison of the mitochondrial genome of three species of Hoplopleura
| 特征 Feature | 缺齿甲胁虱 H. edentula | 克氏甲胁虱 H. kitti | 红姬甲胁虱 H. akanezumi |
|---|---|---|---|
| 微环染色体组成Composition of minichromosome | - | atp8-atp6-N | atp8-atp6-N |
| E-cob-S1(tct)-S2(tga) | E-cob-S1(tct)-S2(tga) | E-cob-S1(tct)-S2(tga) | |
| I-cox1 | I-cox1 | I-cox1 | |
| D-Y-cox2 | D-Y-cox2- 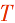 | D-Y-cox2 | |
| - | R-nad4L-P-cox3-A | R-nad4L-P-cox3-A- 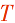 | |
| Q-nad1-G-nad3 | Q-nad1-G-nad3 | - | |
| - | nad2 | nad2 | |
| K-nad4 | K-nad4 | K-nad4 | |
H-nad5-F- 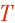 | - | - | |
| - | C-nad6-W-L2(taa) | C-nad6-W-L2(taa) | |
| M-L1(tag)-rrnL-V | M-L1(tag)-rrnL-V | 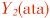 -rrnL-V -rrnL-V | |
| rrnS | rrnS | rrnS | |
| 微环染色 体数No. minichromosome | 8 | 11 | 11 |
| 基因数No. gene | 24 | 34 | 28 |
Table 3
Nucleotide compositions of the mitochondrial genome of H. edentula on E. miletus
| 基因序列 Gene sequence | 长度/bp Length/bp | 占比/% Proportion/% | AT偏倚值 AT-skew | GC偏倚值 GC-skew | ||||
|---|---|---|---|---|---|---|---|---|
| A | T | G | C | A + T | ||||
| 编码区Coding region | 10 545 | 26.9 | 34.1 | 20.7 | 18.3 | 61.0 | -0.118 | 0.062 |
| 蛋白质编码基因 Protein-coding gene | 7 563 | 25.0 | 34.8 | 19.1 | 21.1 | 59.8 | -0.164 | -0.050 |
| 密码子第1位点 The 1st codon position | 2 521 | 28.2 | 28.4 | 25.6 | 17.8 | 56.6 | -0.004 | 0.180 |
| 密码子第2位点 The 2nd codon position | 2 521 | 16.9 | 43.5 | 18.0 | 21.6 | 60.4 | -0.440 | -0.091 |
| 密码子第3位点 The 3rd codon position | 2 521 | 26.8 | 35.7 | 19.5 | 18.0 | 62.5 | -0.426 | 0.040 |
| tRNAs | 995 | 31.5 | 32.8 | 19.6 | 16.1 | 64.3 | -0.020 | 0.098 |
| rrnL | 1 117 | 31.5 | 32.2 | 21.1 | 15.2 | 63.7 | -0.011 | 0.163 |
| rrnS | 874 | 31.2 | 31.5 | 19.2 | 18.1 | 62.7 | -0.005 | 0.029 |
Table 4
Codon usage in protein-coding genes in the mitochondrial genome of H. edentula
| 氨基酸 Amino acid | 密码子 Codon | 使用次数 No. use | 相对同义密码子使用频率 Relative synonymous codon usage | 氨基酸 Amino acid | 密码子 Codon | 使用次数 No. use | 相对同义密码子使用频率 Relative synonymous codon usage |
|---|---|---|---|---|---|---|---|
| 丙氨酸 Ala | GCU | 39 | 1.65 | 脯氨酸 Pro | CCU | 63 | 2.23 |
| GCC | 64 | 1.01 | CCC | 23 | 0.81 | ||
| GCA | 32 | 0.83 | CCA | 16 | 0.57 | ||
| GCG | 20 | 0.52 | CCG | 11 | 0.39 | ||
| 半胱氨酸 Cys | UGU | 21 | 1.08 | 谷氨酰胺 Gln | CAA | 15 | 0.77 |
| UGC | 18 | 0.92 | CAG | 24 | 1.23 | ||
| 天冬氨酸 Asp | GAU | 31 | 1.24 | 精氨酸 Arg | CGU | 6 | 0.27 |
| GAC | 19 | 0.76 | CGC | 10 | 0.45 | ||
| 谷氨酸 Glu | GAA | 41 | 1.32 | CGA | 10 | 0.45 | |
| GAG | 21 | 0.68 | CGG | 12 | 0.54 | ||
| 苯丙氨酸 Phe | UUU | 129 | 1.50 | AGA | 43 | 1.93 | |
| UUC | 43 | 0.50 | AGG | 53 | 2.37 | ||
| 甘氨酸 Gly | GGU | 24 | 0.57 | 丝氨酸 Ser | UCU | 77 | 2.23 |
| GGC | 25 | 0.60 | UCC | 40 | 1.16 | ||
| GGA | 50 | 1.20 | UCA | 40 | 1.16 | ||
| GGG | 68 | 1.63 | UCG | 12 | 0.35 | ||
| 组氨酸 His | CAU | 24 | 0.92 | AGU | 21 | 0.61 | |
| CAC | 28 | 1.08 | AGC | 17 | 0.49 | ||
| 异亮氨酸 Ile | AUU | 137 | 1.53 | 苏氨酸 Thr | ACU | 46 | 1.72 |
| AUC | 38 | 0.43 | ACC | 26 | 0.97 | ||
| AUA | 93 | 1.04 | ACA | 30 | 1.12 | ||
| 赖氨酸 Lys | AAA | 25 | 0.82 | ACG | 5 | 0.19 | |
| AAG | 36 | 1.18 | 缬氨酸 Val | GUU | 75 | 1.42 | |
| 亮氨酸 Leu | UUA | 124 | 1.97 | GUC | 32 | 0.60 | |
| UUG | 46 | 0.73 | GUA | 65 | 1.23 | ||
| CUU | 91 | 1.45 | GUG | 40 | 0.75 | ||
| CUC | 32 | 0.51 | 色氨酸 Trp | UGG | 32 | 1.00 | |
| CUA | 41 | 0.65 | UGA | 45 | 2.60 | ||
| CUG | 43 | 0.68 | 酪氨酸 Tyr | UAU | 46 | 1.12 | |
| 蛋氨酸 Met | AUG | 68 | 1.00 | UAC | 36 | 0.88 | |
| 天冬酰胺 Asn | AAU | 45 | 1.25 | 终止密码子* Stop codon* | UAA | 6 | 0.35 |
| AAC | 27 | 0.75 | UAG | 1 | 0.06 |
| [1] | Durden LA,, Musser GG. The sucking lice (Insecta, Anoplura) of the world: a taxonomic checklist with records of mammalian hosts and geographical distributions[J]. Bull Am Mus Nat Hist, 1994, 218: 1-90. |
| [2] | Chin TH. Taxonomy and fauna of sucking lice (Anoplura) in China[M]. Beijing: Science Press, 1999: 44-45. (in Chinese) |
| (金大雄. 中国吸虱的分类与检索[M]. 北京: 科学出版社, 1999: 44-45.) | |
| [3] | Luo ZX,, Chen W,, Gao W. Fauna sinica[M]. Beijing: Science Press, 2000: 449-451. (in Chinese) |
| (罗泽珣,, 陈卫,, 高武. 中国动物志[M]. 北京: 科学出版社, 2000: 449-451.) | |
| [4] | Zhu WL,, Jia T,, Lian X, et al. Seasonal variations of maximum metabolic rate in Eothenomys miletusin Hengduan mountains region[J]. Acta Ecol Sin, 2010, 30(5): 1133-1139. (in Chinese) |
| (朱万龙,, 贾婷,, 练硝, 等. 横断山脉大绒鼠最大代谢率的季节性差异[J]. 生态学报, 2010, 30(5): 1133-1139.) | |
| [5] | Guo M,, Dong XQ. Development situation of the plague foci Apodemus chevrieri and Eothenomys miletus in northwest Yunnan Province[J]. Chin J Control of Endem Dis, 2008, 23(1): 27-31. (in Chinese) |
| (郭牧,, 董兴齐. 滇西北齐氏姬鼠、大绒鼠鼠疫疫源地的发展概况[J]. 中国地方病防治杂志, 2008, 23(1): 27-31.) | |
| [6] | Dong WG,, Guo XG,, Men XY, et al. Ectoparasites of Eothenomys miletus in the focus of plague in northwest Yunnan[J]. Sichuan J Zool, 2009, 28(5): 683-690. |
| [7] | Peng PY,, Guo XG,, Song WY, et al. Investigation of ectoparasites on body surface of Eothenomys miletus in some places of Guizhou[J]. Guizhou Agric Sci, 2015, 43(2): 75-79. (in Chinese) |
| (彭培英,, 郭宪国,, 宋文宇, 等. 贵州省部分地区大绒鼠体表寄生虫调查[J]. 贵州农业科学, 2015, 43(2): 75-79.) | |
| [8] | Zhang YZ,, Zhang HL,, Mi ZQ, et al. Monitoring of hemorrhagic fever with renal syndrome in Yunnan Province, China, 2005[J]. Chin J Vector Biol Control, 2008, 19(2): 148-150. (in Chinese) |
| (张云智,, 张海林,, 米竹青, 等. 2005年云南省肾综合征出血热监测研究[J]. 中国媒介生物学及控制杂志, 2008, 19(2): 148-150.) | |
| [9] |
Boore JL. Animal mitochondrial genomes[J]. Nucleic Acids Res, 1999, 27(8): 1767-1780.
pmid: 10101183 |
| [10] |
Cameron SL. Insect mitochondrial genomics: implications for evolution and phylogeny[J]. Annu Rev Entomol, 2014, 59(1): 95-117.
doi: 10.1146/annurev-ento-011613-162007 |
| [11] |
Jiang H,, Barker SC,, Shao R. Substantial variation in the extent of mitochondrial genome fragmentation among blood-sucking lice of mammals[J]. Genome Biol Evol, 2013, 5(7): 1298-1308.
doi: 10.1093/gbe/evt094 |
| [12] |
Song SD,, Barker SC,, Shao R. Variation in mitochondrial minichromosome composition between blood-sucking lice of the genus Haematopinus that infest horses and pigs[J]. Parasit Vectors, 2014, 7: 144.
doi: 10.1186/1756-3305-7-144 |
| [13] |
Shao R,, Kirkness EF,, Barker SC. The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus[J]. Genome Res, 2009, 19(5): 904-912.
doi: 10.1101/gr.083188.108 |
| [14] |
Shao R,, Zhu XQ,, Barker SC, et al. Evolution of extensively fragmented mitochondrial genomes in the lice of humans[J]. Genome Biol Evol, 2012, 4(11): 1088-1101.
doi: 10.1093/gbe/evs088 |
| [15] |
Dong WG,, Song S,, Guo XG, et al. Fragmented mitochondrial genomes are present in both major clades of the blood-sucking lice(Suborder Anoplura): evidence from two Hoplopleura rodent lice (family Hoplopleuridae)[J]. BMC Genom, 2014, 15(1): 1-13.
doi: 10.1186/1471-2164-15-1 |
| [16] |
Dong WG,, Song S,, Jin DC, et al. Fragmented mitochondrial genomes of the rat lice, Polyplax asiatica and Polyplax spinulosa: intra-genus variation in fragmentation pattern and a possible link between the extent of fragmentation and the length of life cycle[J]. BMC Genom, 2014, 15: 44.
doi: 10.1186/1471-2164-15-44 |
| [17] |
Shao R,, Li H,, Barker SC, et al. The mitochondrial genome of the guanaco louse, Microthoracius praelongiceps: insights into the ancestral mitochondrial karyotype of sucking lice (Anoplura, Insecta)[J]. Genome Biol Evol, 2017, 9(2): 431-445.
doi: 10.1093/gbe/evx007 |
| [18] |
Fu YT,, Dong Y,, Wang W, et al. Fragmented mitochondrial genomes evolved in opposite directions between closely related macaque louse Pedicinus obtusus and colobus louse Pedicinus badii[J]. Genomics, 2020, 112(6): 4924-4933.
doi: 10.1016/j.ygeno.2020.09.005 |
| [19] |
Herd KE,, Barker SC,, Shao R. The mitochondrial genome of the chimpanzee louse, Pediculus schaeffi: insights into the process of mitochondrial genome fragmentation in the blood-sucking lice of great apes[J]. BMC Genom, 2015, 16: 661.
doi: 10.1186/s12864-015-1843-3 |
| [20] |
Kearse M,, Moir R,, Wilson A, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data[J]. Bioinformatics, 2012, 28(12): 1647-1649.
doi: 10.1093/bioinformatics/bts199 |
| [21] |
Laslett D,, Canbäck B. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences[J]. Bioinformatics, 2008, 24(2): 172-175.
pmid: 18033792 |
| [22] |
Lowe TM,, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence[J]. Nucleic Acids Res, 1997, 25(5): 955-964.
pmid: 9023104 |
| [23] |
Altschul SF,, Madden TL,, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs[J]. Nucleic Acids Res, 1997, 25(17): 3389-3402.
doi: 10.1093/nar/25.17.3389 pmid: 9254694 |
| [24] |
Gish W,, States DJ. Identification of protein coding regions by database similarity search[J]. Nat Genet, 1993, 3(3): 266-272.
pmid: 8485583 |
| [25] |
Puigbò P,, Bravo IG,, Garcia-Vallve S. CAIcal: a combined set of tools to assess codon usage adaptation[J]. Biol Direct, 2008, 3: 38.
doi: 10.1186/1745-6150-3-38 pmid: 18796141 |
| [26] |
Perna NT,, Kocher TD. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes[J]. J Mol Evol, 1995, 41(3): 353-358.
pmid: 7563121 |
| [27] |
Shao R,, Barker SC,, Li H, et al. Fragmented mitochondrial genomes in two suborders of parasitic lice of eutherian mammals (Anoplura and Rhynchophthirina, Insecta)[J]. Sci Rep, 2015, 5: 17389.
doi: 10.1038/srep17389 |
| [28] |
Song F,, Li H,, Liu GH, et al. Mitochondrial genome fragmentation unites the parasitic lice of eutherian mammals[J]. Syst Biol, 2019, 68(3): 430-440.
doi: 10.1093/sysbio/syy062 pmid: 30239978 |
| [29] |
Shi GH,, Cui ZX,, Hui M, et al. The complete mitochondrial genomes of Umalia orientalis and Lyreidus brevifrons: the phylogenetic position of the family Raninidae within Brachyuran crabs[J]. Mar Genom, 2015, 21: 53-61.
doi: 10.1016/j.margen.2015.02.002 |
| [30] | Chen XX,, Yuan ZW,, Yuan XW, et al. Advances in mitochondrial genome complete sequence structure of leafhopper[J]. Genom Appl Biol, 2020, 39(6): 2565-2577. (in Chinese) |
| (陈晓晓,, 袁周伟,, 苑晓伟, 等. 叶蝉线粒体基因组全序列结构研究进展[J]. 基因组学与应用生物学, 2020, 39(6): 2565-2577.) | |
| [31] | Kolesnikov AA,, Gerasimov ES. Diversity of mitochondrial genome organization[J]. Biochemistry(Mosc), 2012, 77(13): 1424-1435. |
| [32] | Li JJ. Research on phylogeny and acoustic signal evolution of Ensifera based on complete mitochondrial genome[D]. Chang-chun:Northeast Normal University, 2018: 13-28. (in Chinese) |
| (李君健. 基于线粒体基因组的螽亚目昆虫系统发育与鸣声进化研究[D]. 长春:东北师范大学, 2018: 13-28.) | |
| [33] | Ou J. Complete mitochondrial genomes sequences of six stored grain beetles[D]. Xi’an:Shanxi Normal University, 2015: 15-57. (in Chinese) |
| (欧静. 六种储粮甲虫线粒体基因组测定及分析[D]. 西安:陕西师范大学, 2015: 15-57.) | |
| [34] | Zhou F. Complete mitochondrial genomes sequences of four locust and analyzed the phylogeny of Orthoptera[D]. Xi’an:Shanxi Normal University, 2015: 27-50. (in Chinese) |
| (周飞. 四种蝗虫线粒体基因组测定及直翅目系统发生分析[D]. 西安:陕西师范大学, 2015: 27-50.) | |
| [35] | Wu YM. Genome sequencing and transcriptome analysis of chemosensory genes of two meloids[D]. Guiyang:Guizhou University, 2018: 36-41. (in Chinese) |
| (吴渊明. 两种芫菁的全基因组测序和化学感受器转录组基因的鉴定与分析[D]. 贵阳:贵州大学, 2018: 36-41.) | |
| [36] | Xia LY,, Sun WW,, Cui M, et al. Sequencing and analysis of mitochondrial genome of Lasioderma serricorne[J]. Tob Sci Technol, 2020, 53(12): 1-8. (in Chinese) |
| (夏丽媛,, 孙为伟,, 崔淼, 等. 烟草甲线粒体基因组序列测定与分析[J]. 烟草科技, 2020, 53(12): 1-8.) | |
| [37] | Liu J,, Bian X. Characteristics of the Orthoptera mitogenome and its application[J]. J Guangxi Norm Univ Nat Sci Ed, 2021, 39(1): 17-28. (in Chinese) |
| 刘静,, 边迅. 直翅目昆虫线粒体基因组的特征及应用[J]. 广西师范大学学报(自然科学版), 2021, 39(1): 17-28.) | |
| [38] |
Yan D,, Tang Y,, Hu M, et al. The mitochondrial genome of Frankliniella intonsa: insights into the evolutionof mitochondrial genomes at lower taxonomic levels in Thysanoptera[J]. Genomics, 2014, 104(4): 306-312.
doi: 10.1016/j.ygeno.2014.08.003 |
| [39] | Chen S. Complete mitochondrial genomes sequences of six species of Spilomelinae[D]. Xi’an: Shanxi Normal University, 2017: 34-37. (in Chinese) |
| (陈汕. 六种斑野螟亚科昆虫全线粒体基因组的序列测定与分析[D]. 西安:陕西师范大学, 2017: 34-37.) | |
| [40] |
Varani G,, Mcclain WH. The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems[J]. Embo Rep, 2000, 1(1): 18-23.
pmid: 11256617 |
| [41] | Wang XM. Structural characteristics of mitochondrial genome and preliminary phylogenetic analysis for Lepus hainanus[D]. Jinan: Shandong University, 2012: 3-4. (in Chinese) |
| (王晓明. 海南兔的线粒体基因组结构特征与系统演化初探[D]. 济南:山东大学, 2012: 3-4.) | |
| [42] |
Broughton RE,, Dowling TE. Length variation in mitochondrial DNA of the minnow Cyprinella spiloptera[J]. Genetics, 1994, 138(1): 179-190.
doi: 10.1093/genetics/138.1.179 pmid: 8001785 |
| [43] |
Ladoukakis ED,, Zouros E. Direct evidence for homologous recombination in mussel (Mytilus galloprovincialis) mitochondrial DNA[J]. Mol Biol Evol, 2001, 18(7): 1168-1175.
pmid: 11420358 |
| [44] |
Okada K,, Yamazaki Y,, Yokobori S, et al. Repetitive sequences in the lamprey mitochondrial DNA control region and speciation of Lethenteron[J]. Gene, 2010, 465(1/2): 45-52.
doi: 10.1016/j.gene.2010.06.009 |
| [45] |
Ray DA,, Densmore LD. Repetitive sequences in the crocodilian mitochondrial control region: Poly-A sequences and heteroplasmic tandem repeats[J]. Mol Biol Evol, 2003, 20(6): 1006-1013.
doi: 10.1093/molbev/msg117 |
| [46] |
Shi W,, Kong XY,, Wang ZM, et al. Pause-melting misalignment: a novel model for the birth and motif indel of tandem repeats in the mitochondrial genome[J]. BMC Genom, 2013, 14: 103.
doi: 10.1186/1471-2164-14-103 |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||